Fluorescence-based investigations of RNA-small molecule interactions
- PMID: 31129289
- PMCID: PMC6756994
- DOI: 10.1016/j.ymeth.2019.05.017
Fluorescence-based investigations of RNA-small molecule interactions
Abstract
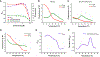
Interrogating non-coding RNA structures and functions with small molecules is an area of rapidly increasing interest among biochemists and chemical biologists. However, many biochemical approaches to monitoring RNA structures are time-consuming and low-throughput, and thereby are only of limited utility for RNA-small molecule studies. Fluorescence-based techniques are powerful tools for rapid investigation of RNA conformations, dynamics, and interactions with small molecules. Many fluorescence methods are amenable to high-throughput analysis, enabling library screening for small molecule binders. In this review, we summarize numerous fluorescence-based approaches for identifying and characterizing RNA-small molecule interactions. We describe in detail a high-information content dual-reporter FRET assay we developed to characterize small molecule-induced conformational and stability changes. Our assay is uniquely suited as a platform for both small molecule discovery and thorough characterization of RNA-small molecule binding mechanisms. Given the growing recognition of non-coding RNAs as attractive targets for therapeutic intervention, we anticipate our FRET assay and other fluorescence-based techniques will be indispensable for the development of potent and specific small molecule inhibitors targeting RNA.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures



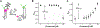

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

