Cyclin F Controls Cell-Cycle Transcriptional Outputs by Directing the Degradation of the Three Activator E2Fs
- PMID: 31130363
- PMCID: PMC6588466
- DOI: 10.1016/j.molcel.2019.04.010
Cyclin F Controls Cell-Cycle Transcriptional Outputs by Directing the Degradation of the Three Activator E2Fs
Abstract
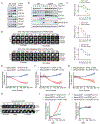
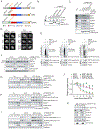
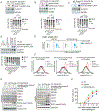
E2F1, E2F2, and E2F3A, the three activators of the E2F family of transcription factors, are key regulators of the G1/S transition, promoting transcription of hundreds of genes critical for cell-cycle progression. We found that during late S and in G2, the degradation of all three activator E2Fs is controlled by cyclin F, the substrate receptor of 1 of 69 human SCF ubiquitin ligase complexes. E2F1, E2F2, and E2F3A interact with the cyclin box of cyclin F via their conserved N-terminal cyclin binding motifs. In the short term, E2F mutants unable to bind cyclin F remain stable throughout the cell cycle, induce unscheduled transcription in G2 and mitosis, and promote faster entry into the next S phase. However, in the long term, they impair cell fitness. We propose that by restricting E2F activity to the S phase, cyclin F controls one of the main and most critical transcriptional engines of the cell cycle.
Keywords: E2F1; E2F2; E2F3A; F-box proteins; RNA-seq; SCF ligases; cell cycle; cyclin F; retinoblastoma; ubiquitin.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
M.P. is a member of the scientific advisory boards of CullGen Inc. and Kymera Therapeutics, and a consultant for BeyondSpring Pharmaceutical.
Figures



References
-
- Bagchi S, Weinmann R, and Raychaudhuri P (1991). The retinoblastoma protein copurifies with E2F-I, an E1A-regulated inhibitor of the transcription factor E2F. Cell 65, 1063–1072. - PubMed
-
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A practical and powerful approach to multiple testing. J Royal Stat Soc 57, 289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

