DiVenn: An Interactive and Integrated Web-Based Visualization Tool for Comparing Gene Lists
- PMID: 31130993
- PMCID: PMC6509638
- DOI: 10.3389/fgene.2019.00421
DiVenn: An Interactive and Integrated Web-Based Visualization Tool for Comparing Gene Lists
Abstract
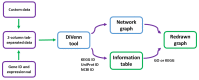
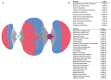
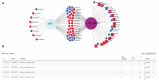
Gene expression data generated from multiple biological samples (mutant, double mutant, and wild-type) are often compared via Venn diagram tools. It is of great interest to know the expression pattern between overlapping genes and their associated gene pathways or gene ontology (GO) terms. We developed DiVenn (Dive into the Venn diagram and create a force directed graph)-a novel web-based tool that compares gene lists from multiple RNA-Seq experiments in a force-directed graph, which shows the gene regulation levels for each gene and integrated KEGG pathway and gene ontology knowledge for the data visualization. DiVenn has four key features: (1) informative force-directed graph with gene expression levels to compare multiple data sets; (2) interactive visualization with biological annotations and integrated pathway and GO databases, which can be used to subset or highlight gene nodes to pathway or GO terms of interest in the graph; (3) Pathway and GO enrichment analysis of all or selected genes in the graph; and (4) high resolution image and gene-associated information export. DiVenn is freely available at http://divenn.noble.org/.
Keywords: KEGG; Venn diagram; gene ontology; pathogen infection; transcriptome data; visualization.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

