CircRNA expression profile of bovine placentas in late gestation with aberrant SCNT fetus
- PMID: 31131498
- PMCID: PMC6642297
- DOI: 10.1002/jcla.22918
CircRNA expression profile of bovine placentas in late gestation with aberrant SCNT fetus
Abstract
Backgrounds: One of the limitations of somatic cell nuclear transfer (SCNT) strategy to generate genetically modified offspring is the low birth rate. Placental dysfunction is one of the causes of abortion. Circular RNA (circRNA) is noncoding RNA which functions as microRNA (miRNA) sponges in biological processes.
Methods: Two aberrant pregnant placenta (aberrant group, AG) and three normal pregnant placenta (normal group, NG) during late gestation (180-210 days) with bovine SCNT fetus were collected for high-throughput sequencing and analyzed. The host genes of differentially expressed (DE) circRNAs were predicted. And the microRNAs (miRNAs) which could interact with DE circRNAs were analyzed. Then, the expressional level of partial DE circRNAs and corresponding host genes was verified through qRT-PCR. At last, the function of host genes was analyzed through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG).
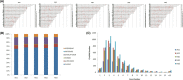
Results: Altogether 123 differentially expressed circRNAs between two groups were identified, which were found related to 60 host genes and 32 miRNAs. The top 10 upregulated circRNAs were bta_circ_0012985, bta_circ_0013071, bta_circ_0013074, bta_circ_0016024, bta_circ_0013068, bta_circ_0008816, bta_circ_0012982, bta_circ_0013072, bta_circ_0019285, and bta_circ_0013067. The top 10 downregulated circRNAs were bta_circ_0024234, bta_circ_0017528, bta_circ_0008077, bta_circ_0003222, bta_circ_0007500, bta_circ_0020328, bta_circ_0011001, bta_circ_0016364, bta_circ_0008839, and bta_circ_0016049. The qRT-PCR results showed consistent trend with sequencing analysis result, while host genes had no statistic difference. The GO and KEGG analyses of the host genes suggested that abnormal circRNA expression may play multiple roles in placental structure and dysfunction.
Conclusion: The abnormal circRNA expression may be one of reasons of placental dysfunction, leads to abortion of bovine SCNT fetus.
Keywords: aberrant development; bovine; circular RNAs; placenta; somatic cell nuclear transfer.
© 2019 The Authors. Journal of Clinical Laboratory Analysis Published by Wiley Periodicals, Inc.
Figures

Similar articles
-
The expression profile of circRNA and its potential regulatory targets in the placentas of severe pre-eclampsia.Taiwan J Obstet Gynecol. 2019 Nov;58(6):769-777. doi: 10.1016/j.tjog.2019.09.007. Taiwan J Obstet Gynecol. 2019. PMID: 31759525
-
Expression profile of circular RNAs in placentas of women with gestational diabetes mellitus.Endocr J. 2019 May 28;66(5):431-441. doi: 10.1507/endocrj.EJ18-0291. Epub 2019 Mar 27. Endocr J. 2019. PMID: 30814439
-
A comprehensive evaluation of skin aging-related circular RNA expression profiles.J Clin Lab Anal. 2021 Apr;35(4):e23714. doi: 10.1002/jcla.23714. Epub 2021 Feb 3. J Clin Lab Anal. 2021. PMID: 33534927 Free PMC article.
-
Studies on the Role of circRNAs in Osteoarthritis.Biomed Res Int. 2021 Sep 4;2021:8231414. doi: 10.1155/2021/8231414. eCollection 2021. Biomed Res Int. 2021. PMID: 34527744 Free PMC article. Review.
-
Circular RNAs: Rising stars in lipid metabolism and lipid disorders.J Cell Physiol. 2021 Jul;236(7):4797-4806. doi: 10.1002/jcp.30200. Epub 2020 Dec 4. J Cell Physiol. 2021. PMID: 33275299 Review.
Cited by
-
Transcription coactivator YAP1 promotes CCND1/CDK6 expression, stimulating cell proliferation in cloned cattle placentas.Zool Res. 2025 Jan 18;46(1):122-138. doi: 10.24272/j.issn.2095-8137.2024.211. Zool Res. 2025. PMID: 39846191 Free PMC article.
-
Genetic Basis of Follicle Development in Dazu Black Goat by Whole-Transcriptome Sequencing.Animals (Basel). 2021 Dec 13;11(12):3536. doi: 10.3390/ani11123536. Animals (Basel). 2021. PMID: 34944311 Free PMC article.
References
-
- Edwards JL, Schrick FN, McCracken MD, et al. Cloning adult farm animals: a review of the possibilities and problems associated with somatic cell nuclear transfer. Am J Reprod Immunol. 2003;50:113‐123. - PubMed
-
- Wells DN. Animal cloning: problems and prospects. Rev Sci Tech. 2005;24:251‐264. - PubMed
-
- Wang F, Kou Z, Zhang Y, Gao S. Dynamic reprogramming of histone acetylation and methylation in the first cell cycle of cloned mouse embryos. Biol Reprod. 2007;77:1007‐1016. - PubMed
-
- Palmieri C, Loi P, Ptak G, Della Salda L. Review paper: a review of the pathology of abnormal placentae of somatic cell nuclear transfer clone pregnancies in cattle, sheep, and mice. Vet Pathol. 2008;45:865‐880. - PubMed
-
- Pozor MA, Sheppard B, Hinrichs K, et al. Placental abnormalities in equine pregnancies generated by SCNT from one donor horse. Theriogenology. 2016;86:1573‐1582. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

