Carbohydrate metabolic systems present on genomic islands are lost and gained in Vibrio parahaemolyticus
- PMID: 31133029
- PMCID: PMC6537148
- DOI: 10.1186/s12866-019-1487-6
Carbohydrate metabolic systems present on genomic islands are lost and gained in Vibrio parahaemolyticus
Abstract
Background: Utilizing unique carbohydrates or utilizing them more efficiently help bacteria expand and colonize new niches. Horizontal gene transfer (HGT) of catabolic systems is a powerful mechanism by which bacteria can acquire new phenotypic traits that can increase survival and fitness in different niches. In this work, we examined carbon catabolism diversity among Vibrio parahaemolyticus, a marine species that is also an important human and fish pathogen.
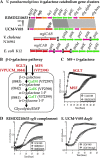
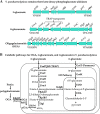
Results: Phenotypic differences in carbon utilization between Vibrio parahaemolyticus strains lead us to examine genotypic differences in this species and the family Vibrionaceae in general. Bioinformatics analysis showed that the ability to utilize D-galactose was present in all V. parahaemolyticus but at least two distinct transporters were present; a major facilitator superfamily (MFS) transporter and a sodium/galactose transporter (SGLT). Growth and genetic analyses demonstrated that SGLT was a more efficient transporter of D-galactose and was the predominant type among strains. Phylogenetic analysis showed that D-galactose gene galM was acquired multiples times within the family Vibrionaceae and was transferred between distantly related species. The ability to utilize D-gluconate was universal within the species. Deletion of eda (VP0065), which encodes aldolase, a key enzyme in the Entner-Doudoroff (ED) pathway, reached a similar biomass to wild type when grown on D-gluconate as a sole carbon source. Two additional eda genes were identified, VPA1708 (eda2) associated with a D-glucuronate cluster and VPA0083 (eda3) that clustered with an oligogalacturonide (OGA) metabolism cluster. EDA2 and EDA3 were variably distributed among the species. A metabolic island was identified that contained citrate fermentation, L-rhamnose and OGA metabolism clusters as well as a CRISPR-Cas system. Phylogenetic analysis showed that CitF and RhaA had a limited distribution among V. parahaemolyticus, and RhaA was acquired at least three times. Within V. parahaemolyticus, two different regions contained the gene for L-arabinose catabolism and most strains had the ability to catabolism this sugar.
Conclusion: Our data suggest that horizontal transfer of metabolic systems among Vibrionaceae is an important source of metabolic diversity. This work identified four EDA homologues suggesting that the ED pathway plays a significant role in metabolism. We describe previously uncharacterized metabolism islands that were hotspots for the gain and loss of functional modules likely mediated by transposons.
Keywords: CRISPR-Cas systems; Citrate fermentation; Entner-Doudoroff aldolase (EDA); L-arabinose; L-rhamnose; Metabolism islands; Sodium/galactose transporter SGLT; Tn7-like transposon.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

