Genomic sequencing of Troides aeacus nucleopolyhedrovirus (TraeNPV) from golden birdwing larvae (Troides aeacus formosanus) to reveal defective Autographa californica NPV genomic features
- PMID: 31133070
- PMCID: PMC6537400
- DOI: 10.1186/s12864-019-5713-2
Genomic sequencing of Troides aeacus nucleopolyhedrovirus (TraeNPV) from golden birdwing larvae (Troides aeacus formosanus) to reveal defective Autographa californica NPV genomic features
Abstract
Background: The golden birdwing butterfly (Troides aeacus formosanus) is a rarely observed species in Taiwan. Recently, a typical symptom of nuclear polyhedrosis was found in reared T. aeacus larvae. From the previous Kimura-2 parameter (K-2-P) analysis based on the nucleotide sequence of three genes in this isolate, polh, lef-8 and lef-9, the underlying virus did not belong to any known nucleopolyhedrovirus (NPV) species. Therefore, this NPV was provisionally named "TraeNPV". To understand this NPV, the nucleotide sequence of the whole TraeNPV genome was determined using next-generation sequencing (NGS) technology.
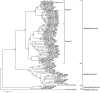
Results: The genome of TraeNPV is 125,477 bp in length with 144 putative open reading frames (ORFs) and its GC content is 40.45%. A phylogenetic analysis based on the 37 baculoviral core genes suggested that TraeNPV is a Group I NPV that is closely related to Autographa californica nucleopolyhedrovirus (AcMNPV). A genome-wide analysis showed that TraeNPV has some different features in its genome compared with other NPVs. Two novel ORFs (Ta75 and Ta139), three truncated ORFs (pcna, he65 and bro) and one duplicated ORF (38.7 K) were found in the TraeNPV genome; moreover, there are fewer homologous regions (hrs) than there are in AcMNPV, which shares eight hrs within the TraeNPV genome. TraeNPV shares similar genomic features with AcMNPV, including the gene content, gene arrangement and gene/genome identity, but TraeNPV lacks 15 homologous ORFs from AcMNPV in its genome, such as ctx, host cell-specific factor 1 (hcf-1), PNK/PNL, vp15, and apsup, which are involved in the auxiliary functions of alphabaculoviruses.
Conclusions: Based on these data, TraeNPV would be clarified as a new NPV species with defective AcMNPV genomic features. The precise relationship between TraeNPV and other closely related NPV species were further investigated. This report could provide comprehensive information on TraeNPV for evolutionary insights into butterfly-infected NPV.
Keywords: TraeNPV; Troides aeacus; Troides aeacus nucleopolyhedrovirus.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
-
- Wu IH, et al. Genetic differentiation of Troides aeacus formosanus (Lepidoptera: Papilionidae), based on cytochrome oxidase I sequences and amplified fragment length polymorphism. Genetics. 2010;130(6):1018–1024.
-
- Haugum J, Low AM. A monograph of the birdwing butterflies, the systematics of Ornithoptera, Troides and related genera: vol. 2, the genera Trogonoptera, Ripponia and Troides. Klampenborg: Scandinavian SciencePress; 1985.
-
- Collins NM, Morris MG. Threatened swallowtail butterßies of the world. Gland and Cambridge: IUCN press; 1985. the IUCN red data book.
-
- Nai Yu-Shin, Huang Yu-Feng, Chen Tzu-Han, Chiu Kuo-Ping, Wang Chung-Hsiung. Biological Control of Pest and Vector Insects. 2017. Determination of Nucleopolyhedrovirus’ Taxonomic Position.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

