High-Diversity Mouse Populations for Complex Traits
- PMID: 31133439
- PMCID: PMC6571031
- DOI: 10.1016/j.tig.2019.04.003
High-Diversity Mouse Populations for Complex Traits
Abstract
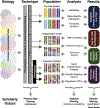
Contemporary mouse genetic reference populations are a powerful platform to discover complex disease mechanisms. Advanced high-diversity mouse populations include the Collaborative Cross (CC) strains, Diversity Outbred (DO) stock, and their isogenic founder strains. When used in systems genetics and integrative genomics analyses, these populations efficiently harnesses known genetic variation for precise and contextualized identification of complex disease mechanisms. Extensive genetic, genomic, and phenotypic data are already available for these high-diversity mouse populations and a growing suite of data analysis tools have been developed to support research on diverse mice. This integrated resource can be used to discover and evaluate disease mechanisms relevant across species.
Keywords: complex genetics; complex traits; genetic diversity; mouse; systems genetics.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Figures

References
-
- Baranov VS et al. (2015) Systems genetics view of endometriosis: A common complex disorder. European Journal of Obstetrics & Gynecology and Reproductive Biology 185, 59–65. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

