The transcriptome of Cryptosporidium oocysts and intracellular stages
- PMID: 31133645
- PMCID: PMC6536522
- DOI: 10.1038/s41598-019-44289-x
The transcriptome of Cryptosporidium oocysts and intracellular stages
Abstract
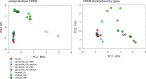
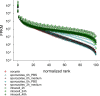
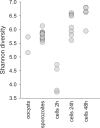
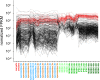
Human cryptosporidiosis is caused primarily by two species of apicomplexan parasites, Cryptosporidium parvum and C. hominis. Although infection of cell monolayers with sporozoites does not support the complete parasite life cycle, the in vitro system is used to study the asexual phase of multiplication, which consists of two generations of merogony. To better understand host-parasite interaction and to gain insight into gene regulatory processes driving the complex life cycle of Cryptosporidium parasites, we analyzed the transcriptome of C. parvum in oocysts, sporozoites and infected cell monolayers 2-48 h post-infection. Analysis of RNA-Seq data from replicate oocyst, sporozoite and intracellular samples revealed significant differences between transcriptomes expressed outside and inside the host cell. Compared to the transcriptome found in the host cell, the oocyst transcriptome is less diverse. Biological processes significantly over-represented intracellularly relate to biosynthetic processes. Genes significantly overexpressed in oocysts show evidence of specialized functions not found in other Apicomplexa. A more comprehensive view of gene regulation during the Cryptosporidium life cycle will require the analysis of later time points during the infection, particularly of the poorly studied sexual phase of the life cycle.
Conflict of interest statement
The authors declare no competing interests.
Figures