Transcriptional and metabolic rewiring of colorectal cancer cells expressing the oncogenic KRASG13D mutation
- PMID: 31133691
- PMCID: PMC6738113
- DOI: 10.1038/s41416-019-0477-7
Transcriptional and metabolic rewiring of colorectal cancer cells expressing the oncogenic KRASG13D mutation
Abstract
Background: Activating mutations in KRAS frequently occur in colorectal cancer (CRC) patients, leading to resistance to EGFR-targeted therapies.
Methods: To better understand the cellular reprogramming which occurs in mutant KRAS cells, we have undertaken a systems-level analysis of four CRC cell lines which express either wild type (wt) KRAS or the oncogenic KRASG13D allele (mtKRAS).
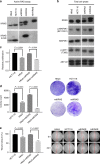
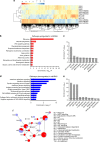
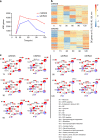
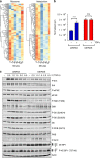
Results: RNAseq revealed that genes involved in ribosome biogenesis, mRNA translation and metabolism were significantly upregulated in mtKRAS cells. Consistent with the transcriptional data, protein synthesis and cell proliferation were significantly higher in the mtKRAS cells. Targeted metabolomics analysis also confirmed the metabolic reprogramming in mtKRAS cells. Interestingly, mtKRAS cells were highly transcriptionally responsive to EGFR activation by TGFα stimulation, which was associated with an unexpected downregulation of genes involved in a range of anabolic processes. While TGFα treatment strongly activated protein synthesis in wtKRAS cells, protein synthesis was not activated above basal levels in the TGFα-treated mtKRAS cells. This was likely due to the defective activation of the mTORC1 and other pathways by TGFα in mtKRAS cells, which was associated with impaired activation of PKB signalling and a transient induction of AMPK signalling.
Conclusions: We have found that mtKRAS cells are substantially rewired at the transcriptional, translational and metabolic levels and that this rewiring may reveal new vulnerabilities in oncogenic KRAS CRC cells that could be exploited in future.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

