AIF-regulated oxidative phosphorylation supports lung cancer development
- PMID: 31133695
- PMCID: PMC6796841
- DOI: 10.1038/s41422-019-0181-4
AIF-regulated oxidative phosphorylation supports lung cancer development
Abstract
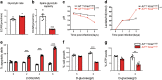
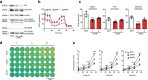
Cancer is a major and still increasing cause of death in humans. Most cancer cells have a fundamentally different metabolic profile from that of normal tissue. This shift away from mitochondrial ATP synthesis via oxidative phosphorylation towards a high rate of glycolysis, termed Warburg effect, has long been recognized as a paradigmatic hallmark of cancer, supporting the increased biosynthetic demands of tumor cells. Here we show that deletion of apoptosis-inducing factor (AIF) in a KrasG12D-driven mouse lung cancer model resulted in a marked survival advantage, with delayed tumor onset and decreased malignant progression. Mechanistically, Aif deletion leads to oxidative phosphorylation (OXPHOS) deficiency and a switch in cellular metabolism towards glycolysis in non-transformed pneumocytes and at early stages of tumor development. Paradoxically, although Aif-deficient cells exhibited a metabolic Warburg profile, this bioenergetic change resulted in a growth disadvantage of KrasG12D-driven as well as Kras wild-type lung cancer cells. Cell-autonomous re-expression of both wild-type and mutant AIF (displaying an intact mitochondrial, but abrogated apoptotic function) in Aif-knockout KrasG12D mice restored OXPHOS and reduced animal survival to the same level as AIF wild-type mice. In patients with non-small cell lung cancer, high AIF expression was associated with poor prognosis. These data show that AIF-regulated mitochondrial respiration and OXPHOS drive the progression of lung cancer.
Conflict of interest statement
The authors declare no competing interests.
Figures






Comment in
-
AIF: an acquired metabolic liability in lung cancer.Cell Res. 2019 Aug;29(8):607-608. doi: 10.1038/s41422-019-0197-9. Cell Res. 2019. PMID: 31267018 Free PMC article. No abstract available.
References
-
- Susin S, et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. - PubMed
-
- Vanessa PM, et al. Downregulation of apoptosis-inducing factor in Harlequin mutant mice sensitizes the myocardium to oxidative stress-related cell death and pressure overload-induced decompensation. Circ. Res. 2005;96:e92–e101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

