Atlas of group A streptococcal vaccine candidates compiled using large-scale comparative genomics
- PMID: 31133745
- PMCID: PMC6650292
- DOI: 10.1038/s41588-019-0417-8
Atlas of group A streptococcal vaccine candidates compiled using large-scale comparative genomics
Erratum in
-
Author Correction: Atlas of group A streptococcal vaccine candidates compiled using large-scale comparative genomics.Nat Genet. 2019 Aug;51(8):1295. doi: 10.1038/s41588-019-0482-z. Nat Genet. 2019. PMID: 31324894
Abstract
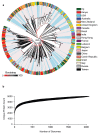
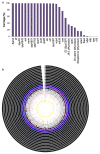
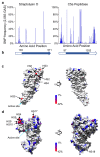
Group A Streptococcus (GAS; Streptococcus pyogenes) is a bacterial pathogen for which a commercial vaccine for humans is not available. Employing the advantages of high-throughput DNA sequencing technology to vaccine design, we have analyzed 2,083 globally sampled GAS genomes. The global GAS population structure reveals extensive genomic heterogeneity driven by homologous recombination and overlaid with high levels of accessory gene plasticity. We identified the existence of more than 290 clinically associated genomic phylogroups across 22 countries, highlighting challenges in designing vaccines of global utility. To determine vaccine candidate coverage, we investigated all of the previously described GAS candidate antigens for gene carriage and gene sequence heterogeneity. Only 15 of 28 vaccine antigen candidates were found to have both low naturally occurring sequence variation and high (>99%) coverage across this diverse GAS population. This technological platform for vaccine coverage determination is equally applicable to prospective GAS vaccine antigens identified in future studies.
Conflict of interest statement
AS is an employee of the GSK group of companies having a commercial interest in GAS vaccine development. The company had no influence over study design.
Figures
References
-
- Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–94. - PubMed
-
- Watkins DA, et al. Global, regional, and national burden of rheumatic heart disease, 1990-2015. N Engl J Med. 2017;377:713–722. - PubMed
-
- Henningham A, Gillen CM, Walker MJ. Group A streptococcal vaccine candidates: potential for the development of a human vaccine. Curr Top Microbiol Immunol. 2013;368:207–42. - PubMed
-
- Kotloff KL, et al. Safety and immunogenicity of a recombinant multivalent group A streptococcal vaccine in healthy adults: phase 1 trial. JAMA. 2004;292:709–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

