Structural basis for neutralization of Plasmodium vivax by naturally acquired human antibodies that target DBP
- PMID: 31133752
- PMCID: PMC6707876
- DOI: 10.1038/s41564-019-0461-2
Structural basis for neutralization of Plasmodium vivax by naturally acquired human antibodies that target DBP
Erratum in
-
Author Correction: Structural basis for neutralization of Plasmodium vivax by naturally acquired human antibodies that target DBP.Nat Microbiol. 2019 Nov;4(11):2024. doi: 10.1038/s41564-019-0587-2. Nat Microbiol. 2019. PMID: 31551566
Abstract
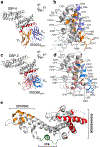
The Plasmodium vivax Duffy-binding protein (DBP) is a prime target of the protective immune response and a promising vaccine candidate for P. vivax malaria. Naturally acquired immunity (NAI) protects against malaria in adults residing in infection-endemic regions, and the passive transfer of malarial immunity confers protection. A vaccine that replicates NAI will effectively prevent disease. Here, we report the structures of DBP region II in complex with human-derived, neutralizing monoclonal antibodies obtained from an individual in a malaria-endemic area with NAI. We identified protective epitopes using X-ray crystallography, hydrogen-deuterium exchange mass spectrometry, mutational mapping and P. vivax invasion studies. These approaches reveal that naturally acquired human antibodies neutralize P. vivax by targeting the binding site for Duffy antigen receptor for chemokines (DARC) and the dimer interface of P. vivax DBP. Antibody binding is unaffected by polymorphisms in the vicinity of epitopes, suggesting that the antibodies have evolved to engage multiple polymorphic variants of DBP. The human antibody epitopes are broadly conserved and are distinct from previously defined epitopes for broadly conserved murine monoclonal antibodies. A library of globally conserved epitopes of neutralizing human antibodies offers possibilities for rational design of strain-transcending DBP-based vaccines and therapeutics against P. vivax.
Conflict of interest statement
COMPETING INTERESTS
The authors declare no competing interests.
Figures



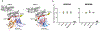

Comment in
-
Structure Solves the Problem with Malaria Merozoite Vaccines.Trends Parasitol. 2019 Nov;35(11):855-857. doi: 10.1016/j.pt.2019.09.004. Epub 2019 Oct 14. Trends Parasitol. 2019. PMID: 31623952 Free PMC article.
References
-
- Cole-Tobian JL et al. Age-acquired immunity to a Plasmodium vivax invasion ligand, the duffy binding protein. J Infect Dis 186, 531–9 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

