Probiotic Lactobacillus rhamnosus GG Induces Alterations in Ileal Microbiota With Associated CD3-CD19-T-bet+IFNγ+/- Cell Subset Homeostasis in Pigs Challenged With Salmonella enterica Serovar 4,[5],12:i:
- PMID: 31134022
- PMCID: PMC6516042
- DOI: 10.3389/fmicb.2019.00977
Probiotic Lactobacillus rhamnosus GG Induces Alterations in Ileal Microbiota With Associated CD3-CD19-T-bet+IFNγ+/- Cell Subset Homeostasis in Pigs Challenged With Salmonella enterica Serovar 4,[5],12:i:
Abstract
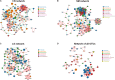
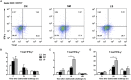
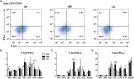
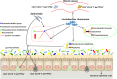
Salmonella enterica serovar 4,[5],12:i:- (S. 4,[5],12:i:-) is an emerging foodborne pathogen causing salmonellosis in humans and animals. Probiotic Lactobacillus rhamnosus GG (LGG) is an effective strategy for controlling enteric infections through maintaining gut microbiota homeostasis and regulating the intestinal innate immune response. Here, LGG was orally administrated to newly weaned piglets for 1 week before S. 4,[5],12:i:- challenge. S. 4,[5],12:i:- challenge led to disturbed gut microbiota, characterized by increased levels of Psychrobacter, Chryseobacterium indoltheticum, and uncultured Corynebacteriaceae populations, as well as an aberrant correlation network in Prevotellaceae NK3B31 group-centric species. The beneficial effect of LGG correlated with attenuating the expansion of Prevotellaceae NK3B31 group. Fusobacterium only found in the pigs treated with LGG was positively correlated with Lactobacillus animalis and Propionibacterium. Administration of LGG induced the expansion of CD3-CD19-T-bet+IFNγ+ and CD3-CD19-T-bet+IFNγ- cell subsets in the peripheral blood at 24 h after a challenge of S. 4,[5],12:i:-. S. 4,[5],12:i:- infection increased the population of intraepithelial CD3-CD19-T-bet+IFNγ+ and CD3-CD19-T-bet+IFNγ- cells in the ileum; however, this increase was attenuated via LGG administration. Correlation analysis revealed that LGG enriched Flavobacterium frigidarium and Facklamia populations, which were negatively correlated with intraepithelial CD3-CD19-T-bet+IFNγ+ and CD3-CD19-T-bet+IFNγ- cells in the ileum. The present data suggest that probiotic LGG alters gut microbiota with associated CD3-CD19-T-bet+IFNγ+/- cell subset homeostasis in pigs challenged with S. enterica 4,[5],12:i:-. LGG may be used in potential gut microbiota-targeted therapy regimens to regulate the specific immune cell function and, consequently, control enteric infections.
Keywords: 12:i:-; IFNγ; Lactobacillus rhamnosus; Salmonella enterica serovar 4; T-bet; [5]; gut microbiota; pig.
Figures



Similar articles
-
Oral Administration of Lactobacillus rhamnosus GG Ameliorates Salmonella Infantis-Induced Inflammation in a Pig Model via Activation of the IL-22BP/IL-22/STAT3 Pathway.Front Cell Infect Microbiol. 2017 Jul 18;7:323. doi: 10.3389/fcimb.2017.00323. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28770173 Free PMC article.
-
Lactobacillus rhamnosus GG Affects Microbiota and Suppresses Autophagy in the Intestines of Pigs Challenged with Salmonella Infantis.Front Microbiol. 2018 Jan 17;8:2705. doi: 10.3389/fmicb.2017.02705. eCollection 2017. Front Microbiol. 2018. PMID: 29403451 Free PMC article.
-
Anti-inflammatory capacity of Lactobacillus rhamnosus GG in monophasic variant Salmonella infected piglets is correlated with impeding NLRP6-mediated host inflammatory responses.Vet Microbiol. 2017 Oct;210:91-100. doi: 10.1016/j.vetmic.2017.08.008. Epub 2017 Aug 20. Vet Microbiol. 2017. PMID: 29103703
-
Gut Microbiota and Cancer: From Pathogenesis to Therapy.Cancers (Basel). 2019 Jan 3;11(1):38. doi: 10.3390/cancers11010038. Cancers (Basel). 2019. PMID: 30609850 Free PMC article. Review.
-
Research progress on the application of Lacticaseibacillus rhamnosus GG in pediatric respiratory diseases.Front Nutr. 2025 Feb 21;12:1553674. doi: 10.3389/fnut.2025.1553674. eCollection 2025. Front Nutr. 2025. PMID: 40062233 Free PMC article. Review.
Cited by
-
Maternal probiotic mixture supplementation optimizes the gut microbiota structure of offspring piglets through the gut-breast axis.Anim Nutr. 2024 Jul 17;19:386-400. doi: 10.1016/j.aninu.2024.04.025. eCollection 2024 Dec. Anim Nutr. 2024. PMID: 39640549 Free PMC article.
-
Temporal and nutritional effects on the weaner pig ileal microbiota.Anim Microbiome. 2021 Aug 28;3(1):58. doi: 10.1186/s42523-021-00119-y. Anim Microbiome. 2021. PMID: 34454628 Free PMC article.
-
Beneficial roles of probiotics on the modulation of gut microbiota and immune response in pigs.PLoS One. 2019 Aug 28;14(8):e0220843. doi: 10.1371/journal.pone.0220843. eCollection 2019. PLoS One. 2019. PMID: 31461453 Free PMC article.
-
Mechanisms and applications of probiotics in prevention and treatment of swine diseases.Porcine Health Manag. 2023 Feb 6;9(1):5. doi: 10.1186/s40813-022-00295-6. Porcine Health Manag. 2023. PMID: 36740713 Free PMC article. Review.
-
Transcriptome Analysis Identifies Strategies Targeting Immune Response-Related Pathways to Control Enterotoxigenic Escherichia coli Infection in Porcine Intestinal Epithelial Cells.Front Vet Sci. 2021 Aug 10;8:677897. doi: 10.3389/fvets.2021.677897. eCollection 2021. Front Vet Sci. 2021. PMID: 34447800 Free PMC article.
References
-
- An T. J., Benvenuti M. A., Mignemi M. E., Martus J., Wood J. B., Thomsen I. P., et al. (2017). Similar clinical severity and outcomes for methicillin-resistant and methicillin-susceptible Staphylococcus aureus pediatric musculoskeletal infections. Open Forum Infect. Dis. 4:ofx013. 10.1093/ofid/ofx013 - DOI - PMC - PubMed
-
- Barthel M., Hapfelmeier S., Quintanilla-Martínez L., Kremer M., Rohde M., Hogardt M., et al. (2003). Pretreatment of mice with streptomycin provides a Salmonella enterica serovar typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71 2839–2858. 10.1128/iai.71.5.2839-2858.2003 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

