Rhizobium etli Produces Nitrous Oxide by Coupling the Assimilatory and Denitrification Pathways
- PMID: 31134023
- PMCID: PMC6514139
- DOI: 10.3389/fmicb.2019.00980
Rhizobium etli Produces Nitrous Oxide by Coupling the Assimilatory and Denitrification Pathways
Abstract
More than two-thirds of the powerful greenhouse gas nitrous oxide (N2O) emissions from soils can be attributed to microbial denitrification and nitrification processes. Bacterial denitrification reactions are catalyzed by the periplasmic (Nap) or membrane-bound (Nar) nitrate reductases, nitrite reductases (NirK/cd 1Nir), nitric oxide reductases (cNor, qNor/ CuANor), and nitrous oxide reductase (Nos) encoded by nap/nar, nir, nor and nos genes, respectively. Rhizobium etli CFN42, the microsymbiont of common bean, is unable to respire nitrate under anoxic conditions and to perform a complete denitrification pathway. This bacterium lacks the nap, nar and nos genes but contains genes encoding NirK and cNor. In this work, we demonstrated that R. etli is able to grow with nitrate as the sole nitrogen source under aerobic and microoxic conditions. Genetic and functional characterization of a gene located in the R. etli chromosome and annotated as narB demonstrated that growth under aerobic or microoxic conditions with nitrate as nitrogen source as well as nitrate reductase activity requires NarB. In addition to be involved in nitrate assimilation, NarB is also required for NO and N2O production by NirK and cNor, respectively, in cells grown microoxically with nitrate as the only N source. Furthermore, β-glucuronidase activity from nirK::uidA and norC::uidA fusions, as well as NorC expression and Nir and Nor activities revealed that expression of nor genes under microoxic conditions also depends on nitrate reduction by NarB. Our results suggest that nitrite produced by NarB from assimilatory nitrate reduction is detoxified by NirK and cNor denitrifying enzymes that convert nitrite into NO which in turn is reduced to N2O, respectively.
Keywords: assimilatory nitrate reductase; denitrification; gene expression; nitrous oxide; soil bacteria.
Figures

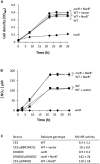
 ), WT + vector (X), narB (
), WT + vector (X), narB ( ), narB + NarB+ (
), narB + NarB+ ( ), and WT + NarB+ (
), and WT + NarB+ ( ) strains were incubated aerobically in minimal medium with NO3− as sole N-source. Data are expressed as the mean value ± SD from two different cultures assayed in triplicate.
) strains were incubated aerobically in minimal medium with NO3− as sole N-source. Data are expressed as the mean value ± SD from two different cultures assayed in triplicate.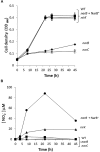
 ), narB (
), narB ( ), narB+ NarB+ (
), narB+ NarB+ ( ), norC (X), and nirK (
), norC (X), and nirK ( ) strains were cultured microoxically in minimal medium with NO3− as sole N-source. Data are expressed as the mean value and error bars from two different cultures assayed in triplicate.
) strains were cultured microoxically in minimal medium with NO3− as sole N-source. Data are expressed as the mean value and error bars from two different cultures assayed in triplicate.

Similar articles
-
Regulation and symbiotic role of nirK and norC expression in Rhizobium etli.Mol Plant Microbe Interact. 2011 Feb;24(2):233-45. doi: 10.1094/MPMI-07-10-0173. Mol Plant Microbe Interact. 2011. PMID: 21043576
-
Effect of Copper on Expression of Functional Genes and Proteins Associated with Bradyrhizobium diazoefficiens Denitrification.Int J Mol Sci. 2022 Mar 21;23(6):3386. doi: 10.3390/ijms23063386. Int J Mol Sci. 2022. PMID: 35328804 Free PMC article.
-
Genetic basis for denitrification in Ensifer meliloti.BMC Microbiol. 2014 Jun 2;14:142. doi: 10.1186/1471-2180-14-142. BMC Microbiol. 2014. PMID: 24888981 Free PMC article.
-
Denitrification in Sinorhizobium meliloti.Biochem Soc Trans. 2011 Dec;39(6):1886-9. doi: 10.1042/BST20110733. Biochem Soc Trans. 2011. PMID: 22103545 Review.
-
Fungal denitrification and nitric oxide reductase cytochrome P450nor.Philos Trans R Soc Lond B Biol Sci. 2012 May 5;367(1593):1186-94. doi: 10.1098/rstb.2011.0335. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22451104 Free PMC article. Review.
Cited by
-
Short-Term Effects of Cenchrus fungigraminus/Potato or Broad Bean Interplanting on Rhizosphere Soil Fertility, Microbial Diversity, and Greenhouse Gas Sequestration in Southeast China.Microorganisms. 2024 Aug 13;12(8):1665. doi: 10.3390/microorganisms12081665. Microorganisms. 2024. PMID: 39203507 Free PMC article.
-
Methane, arsenic, selenium and the origins of the DMSO reductase family.Sci Rep. 2020 Jul 2;10(1):10946. doi: 10.1038/s41598-020-67892-9. Sci Rep. 2020. PMID: 32616801 Free PMC article.
-
Exploring the distribution and co-occurrence of rpf-like genes and nitrogen-cycling genes in water reservoir sediments.Front Microbiol. 2024 Jul 22;15:1433046. doi: 10.3389/fmicb.2024.1433046. eCollection 2024. Front Microbiol. 2024. PMID: 39104579 Free PMC article.
-
An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.Sci Adv. 2021 Jan 1;7(1):eabd8523. doi: 10.1126/sciadv.abd8523. Print 2021 Jan. Sci Adv. 2021. PMID: 33523860 Free PMC article.
-
Divergent profiles of rhizosphere soil carbon and nitrogen cycling in Pinus massoniana provenances with different types of carbon storage.Front Microbiol. 2025 Mar 17;16:1537173. doi: 10.3389/fmicb.2025.1537173. eCollection 2025. Front Microbiol. 2025. PMID: 40165787 Free PMC article.
References
-
- Bedmar E. J., Bueno E., Correa D., Torres M. J., Delgado M. J., Mesa S. (2013). “Ecology of denitrification in soils and plant-associated,” in Beneficial Plant-Microbial Interactions: Ecology and Applications, eds Rodelas B., Gonzalez-López J. (Boca Ratón, FL: CRC Press; ), 164–182.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

