Antibiotic Resistance in Salmonella Typhimurium Isolates Recovered From the Food Chain Through National Antimicrobial Resistance Monitoring System Between 1996 and 2016
- PMID: 31134024
- PMCID: PMC6514237
- DOI: 10.3389/fmicb.2019.00985
Antibiotic Resistance in Salmonella Typhimurium Isolates Recovered From the Food Chain Through National Antimicrobial Resistance Monitoring System Between 1996 and 2016
Erratum in
-
Corrigendum: Antibiotic Resistance in Salmonella Typhimurium Isolates Recovered From the Food Chain Through National Antimicrobial Resistance Monitoring System Between 1996 and 2016.Front Microbiol. 2020 Aug 14;11:1738. doi: 10.3389/fmicb.2020.01738. eCollection 2020. Front Microbiol. 2020. PMID: 32922369 Free PMC article.
Abstract
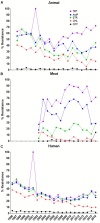
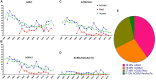
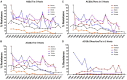
Salmonella is a major foodborne pathogen which causes widespread contamination and infection worldwide. Salmonella Typhimurium is one of the leading serovars responsible for human and animal salmonellosis, globally. The increasing rate of antibiotic resistance in Salmonella Typhimurium poses a significant global concern, and an improved understanding of the distribution of antibiotic resistance patterns in Salmonella Typhimurium is essential for choosing the suitable antibiotic for the treatment of infections. To evaluate the roles of animal and human in antibiotic resistance dissemination, this study aims to categorize 11,447 S. Typhimurium strains obtained across the food-chain, including food animals, retail meats and humans for 21 years in the United States by analyzing minimum inhibitory concentrations (MICs) values for 27 antibiotics. Random Forest Algorithm and Hierarchical Clustering statistics were used to group the strains according to their minimum inhibitory concentration values. Classification and Regression Tree analysis was used to identify the best classifier for human- and animal-populations' isolates. We found the persistent population or multi-drug resistant strains of S. Typhimurium across the four time periods (1996∼2000, 2001∼2005, 2006∼2010, 2011∼2016). Importantly, we also detected that there was more diversity in the MIC patterns among S. Typhimurium strains isolated between 2011 and 2016, which suggests significant emergence of diversified multi-drug resistant strains. The most frequently observed (43%) antibiotic resistance patterns found in S. Typhimurium were tetra-resistant pattern ASSuT (ampicillin, streptomycin, sulfonamides, and tetracycline) and the penta-resistant pattern ACSSuT (ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline). Animals (mainly swine and bovine) are the major source for these two frequently found antibiotic resistance patterns. The occurrence of antibiotic resistant strains from humans and chicken is alarming. Strains were mostly susceptible to fluoroquinolones. Together, this study helped in understanding the expansion of dynamics of antibiotic resistance of S. Typhimurium and recommended fluoroquinolones as a possible treatment options against S. Typhimurium infection.
Keywords: Salmonella Typhimurium; antibiotic resistance; fluoroquinolones; foodborne pathogen; minimum inhibitory concentration; population diversity.
Figures



References
-
- Adhikari B., Besser T. E., Gay J. M., Fox L. K., Hancock D. D., Davis M. A. (2010). Multilocus variable-number tandem-repeat analysis and plasmid profiling to study the occurrence of blaCMY-2 within a pulsed-field gel electrophoresis-defined clade of Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 76 69–74. 10.1128/AEM.00210-09 - DOI - PMC - PubMed
-
- Anderson A. S., Bauer H., Nelson C. B. (1955). Salmonellosis due to Salmonella typhimurium with Easter chicks as likely source. J. Am. Med. Assoc. 158 1153–1155. - PubMed
LinkOut - more resources
Full Text Sources

