Mucosal and Cutaneous Human Papillomavirus Infections and Cancer Biology
- PMID: 31134154
- PMCID: PMC6517478
- DOI: 10.3389/fonc.2019.00355
Mucosal and Cutaneous Human Papillomavirus Infections and Cancer Biology
Abstract
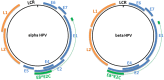
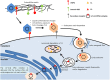
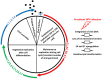
Papillomaviridae is a family of small non-enveloped icosahedral viruses with double-stranded circular DNA. More than 200 different human papillomaviruses (HPVs) have been listed so far. Based on epidemiological data, a subgroup of alphapapillomaviruses (alpha HPVs) was referred to as high-risk (HR) HPV types. HR HPVs are the etiological agents of anogenital cancer and a subset of head and neck cancers. The cutaneous HPV types, mainly from beta and gamma genera, are widely present on the surface of the skin in the general population. However, there is growing evidence of an etiological role of betapapillomaviruses (beta HPVs) in non-melanoma skin cancer (NMSC), together with ultraviolet (UV) radiation. Studies performed on mucosal HR HPV types, such as 16 and 18, showed that both oncoproteins E6 and E7 play a key role in cervical cancer by altering pathways involved in the host immune response to establish a persistent infection and by promoting cellular transformation. Continuous expression of E6 and E7 of mucosal HR HPV types is essential to initiate and to maintain the cellular transformation process, whereas expression of E6 and E7 of cutaneous HPV types is not required for the maintenance of the skin cancer phenotype. Beta HPV types appear to play a role in the initiation of skin carcinogenesis, by exacerbating the accumulation of UV radiation-induced DNA breaks and somatic mutations (the hit-and-run mechanism), and they would therefore act as facilitators rather than direct actors in NMSC. In this review, the natural history of HPV infection and the transforming properties of various HPV genera will be described, with a particular focus on describing the state of knowledge about the role of cutaneous HPV types in NMSC.
Keywords: Papillomavirus; anogenital cancer; infection and cancer; skin cancer; transformation.
Figures
References
-
- Lopez-Bueno A, Mavian C, Labella AM, Castro D, Borrego JJ, Alcami A, et al. . Concurrence of iridovirus, polyomavirus, and a unique member of a new group of fish papillomaviruses in lymphocystis disease-affected gilthead sea bream. J Virol. (2016) 90:8768–79. 10.1128/JVI.01369-16 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

