ExpansionHunter: a sequence-graph-based tool to analyze variation in short tandem repeat regions
- PMID: 31134279
- PMCID: PMC6853681
- DOI: 10.1093/bioinformatics/btz431
ExpansionHunter: a sequence-graph-based tool to analyze variation in short tandem repeat regions
Abstract
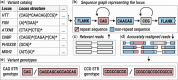
Summary: We describe a novel computational method for genotyping repeats using sequence graphs. This method addresses the long-standing need to accurately genotype medically important loci containing repeats adjacent to other variants or imperfect DNA repeats such as polyalanine repeats. Here we introduce a new version of our repeat genotyping software, ExpansionHunter, that uses this method to perform targeted genotyping of a broad class of such loci.
Availability and implementation: ExpansionHunter is implemented in C++ and is available under the Apache License Version 2.0. The source code, documentation, and Linux/macOS binaries are available at https://github.com/Illumina/ExpansionHunter/.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2019. Published by Oxford University Press.
Figures
References
-
- Amiel J. et al. (2003) Polyalanine expansion and frameshift mutations of the paired-like homeobox gene PHOX2B in congenital central hypoventilation syndrome. Nat. Genet., 33, 459.. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

