The functional false discovery rate with applications to genomics
- PMID: 31135886
- PMCID: PMC7846131
- DOI: 10.1093/biostatistics/kxz010
The functional false discovery rate with applications to genomics
Abstract
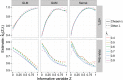
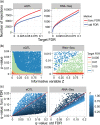
The false discovery rate (FDR) measures the proportion of false discoveries among a set of hypothesis tests called significant. This quantity is typically estimated based on p-values or test statistics. In some scenarios, there is additional information available that may be used to more accurately estimate the FDR. We develop a new framework for formulating and estimating FDRs and q-values when an additional piece of information, which we call an "informative variable", is available. For a given test, the informative variable provides information about the prior probability a null hypothesis is true or the power of that particular test. The FDR is then treated as a function of this informative variable. We consider two applications in genomics. Our first application is a genetics of gene expression (eQTL) experiment in yeast where every genetic marker and gene expression trait pair are tested for associations. The informative variable in this case is the distance between each genetic marker and gene. Our second application is to detect differentially expressed genes in an RNA-seq study carried out in mice. The informative variable in this study is the per-gene read depth. The framework we develop is quite general, and it should be useful in a broad range of scientific applications.
Keywords: q-value; FDR; Functional data analysis; Genetics of gene expression; Kernel density estimation; Local false discovery rate; Multiple hypothesis testing; RNA-seq; Sequencing depth; eQTL.
© The Author 2019. Published by Oxford University Press.
Figures

References
-
- Benjamini, Y. and Heller, R. (2007). False discovery rates for spatial signals. Journal of the American Statistical Association 102, 1272–1281.
-
- Benjamini, Y. and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B 57, 289–300.
-
- Brem, R. B., Yvert, G., Clinton, R. and Kruglyak, L. (2002). Genetic dissection of transcriptional regulation in budding yeast. Science 296, 752–755. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

