The human brain somatostatin interactome: SST binds selectively to P-type family ATPases
- PMID: 31136617
- PMCID: PMC6538167
- DOI: 10.1371/journal.pone.0217392
The human brain somatostatin interactome: SST binds selectively to P-type family ATPases
Abstract
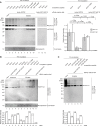
Somatostatin (SST) is a cyclic peptide that is understood to inhibit the release of hormones and neurotransmitters from a variety of cells by binding to one of five canonical G protein-coupled SST receptors (SSTR1 to SSTR5). Recently, SST was also observed to interact with the amyloid beta (Aβ) peptide and affect its aggregation kinetics, raising the possibility that it may bind other brain proteins. Here we report on an SST interactome analysis that made use of human brain extracts as biological source material and incorporated advanced mass spectrometry workflows for the relative quantitation of SST binding proteins. The analysis revealed SST to predominantly bind several members of the P-type family of ATPases. Subsequent validation experiments confirmed an interaction between SST and the sodium-potassium pump (Na+/K+-ATPase) and identified a tryptophan residue within SST as critical for binding. Functional analyses in three different cell lines indicated that SST might negatively modulate the K+ uptake rate of the Na+/K+-ATPase.
Conflict of interest statement
HaW and GS held a provisional patent on amyloid-beta binding polypeptides (filing number 62/451,309) in the year 2017. At this time, the provisional patent expired and no application for a regular patent was filed. The other authors declare that no competing interests exist. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures



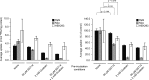
Similar articles
-
Somatostatin in Alzheimer's disease: A new Role for an Old Player.Prion. 2018 Jan 2;12(1):1-8. doi: 10.1080/19336896.2017.1405207. Epub 2018 Jan 31. Prion. 2018. PMID: 29192843 Free PMC article.
-
Somatostatin and its receptor family.Front Neuroendocrinol. 1999 Jul;20(3):157-98. doi: 10.1006/frne.1999.0183. Front Neuroendocrinol. 1999. PMID: 10433861 Review.
-
Molecular cloning, functional characterization, and chromosomal localization of a human somatostatin receptor (somatostatin receptor type 5) with preferential affinity for somatostatin-28.Mol Pharmacol. 1994 Mar;45(3):417-27. Mol Pharmacol. 1994. PMID: 7908405
-
Somatostatin binds to the human amyloid β peptide and favors the formation of distinct oligomers.Elife. 2017 Jun 26;6:e28401. doi: 10.7554/eLife.28401. Elife. 2017. PMID: 28650319 Free PMC article.
-
The somatostatin receptor in human pancreatic β-cells.Vitam Horm. 2014;95:165-93. doi: 10.1016/B978-0-12-800174-5.00007-7. Vitam Horm. 2014. PMID: 24559918 Review.
Cited by
-
Somatostatin and Its Receptors in Myocardial Ischemia/Reperfusion Injury and Cardioprotection.Front Pharmacol. 2021 Nov 5;12:663655. doi: 10.3389/fphar.2021.663655. eCollection 2021. Front Pharmacol. 2021. PMID: 34803662 Free PMC article.
-
Interrogation of the human cortical peptidome uncovers cell-type specific signatures of cognitive resilience against Alzheimer's disease.Sci Rep. 2024 Mar 26;14(1):7161. doi: 10.1038/s41598-024-57104-z. Sci Rep. 2024. PMID: 38531951 Free PMC article.
-
The Role of Receptor-Ligand Interaction in Somatostatin Signaling Pathways: Implications for Neuroendocrine Tumors.Cancers (Basel). 2023 Dec 25;16(1):116. doi: 10.3390/cancers16010116. Cancers (Basel). 2023. PMID: 38201544 Free PMC article. Review.
-
Somatostatin and Its Receptor System in Colorectal Cancer.Biomedicines. 2021 Nov 22;9(11):1743. doi: 10.3390/biomedicines9111743. Biomedicines. 2021. PMID: 34829972 Free PMC article. Review.
-
Wide-Ranging Effects on the Brain Proteome in a Transgenic Mouse Model of Alzheimer's Disease Following Treatment with a Brain-Targeting Somatostatin Peptide.ACS Chem Neurosci. 2021 Jul 7;12(13):2529-2541. doi: 10.1021/acschemneuro.1c00303. Epub 2021 Jun 25. ACS Chem Neurosci. 2021. PMID: 34170117 Free PMC article.
References
-
- Brazeau P, Vale W, Burgus R, Ling N, Butcher M, Rivier J, et al. Hypothalamic polypeptide that inhibits the secretion of immunoreactive pituitary growth hormone. Science. 1973;179(4068):77–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

