Lens differentiation is characterized by stage-specific changes in chromatin accessibility correlating with differentiation state-specific gene expression
- PMID: 31136738
- PMCID: PMC6667291
- DOI: 10.1016/j.ydbio.2019.04.020
Lens differentiation is characterized by stage-specific changes in chromatin accessibility correlating with differentiation state-specific gene expression
Abstract
Changes in chromatin accessibility regulate the expression of multiple genes by controlling transcription factor access to key gene regulatory sequences. Here, we sought to establish a potential function for altered chromatin accessibility in control of key gene expression events during lens cell differentiation by establishing genome-wide chromatin accessibility maps specific for four distinct stages of lens cell differentiation and correlating specific changes in chromatin accessibility with genome-wide changes in gene expression. ATAC sequencing was employed to generate chromatin accessibility profiles that were correlated with the expression profiles of over 10,000 lens genes obtained by high-throughput RNA sequencing at the same stages of lens cell differentiation. Approximately 90,000 regions of the lens genome exhibited distinct changes in chromatin accessibility at one or more stages of lens differentiation. Over 1000 genes exhibited high Pearson correlation coefficients (r > 0.7) between altered expression levels at specific stages of lens cell differentiation and changes in chromatin accessibility in potential promoter (-7.5kbp/+2.5kbp of the transcriptional start site) and/or other potential cis-regulatory regions ( ±10 kb of the gene body). Analysis of these regions identified consensus binding sequences for multiple transcription factors including members of the TEAD, FOX, and NFAT families of transcription factors as well as HIF1a, RBPJ and IRF1. Functional mapping of genes with high correlations between altered chromatin accessibility and differentiation state-specific gene expression changes identified multiple families of proteins whose expression could be regulated through changes in chromatin accessibility including those governing lens structure (BFSP1,BFSP2), gene expression (Pax-6, Sox 2), translation (TDRD7), cell-cell communication (GJA1), autophagy (FYCO1), signal transduction (SMAD3, EPHA2), and lens transparency (CRYBB1, CRYBA4). These data provide a novel relationship between altered chromatin accessibility and lens differentiation and they identify a wide-variety of lens genes and functions that could be regulated through altered chromatin accessibility. The data also point to a large number of potential DNA regulatory sequences and transcription factors whose functional analysis is likely to provide insight into novel regulatory mechanisms governing the lens differentiation program.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures














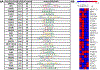






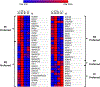
Comment in
-
The developing ocular lens: A practical paradigm for probing chromatin landscape and gene expression.Dev Biol. 2019 Sep 1;453(1):105-106. doi: 10.1016/j.ydbio.2019.05.010. Epub 2019 May 23. Dev Biol. 2019. PMID: 31129080 No abstract available.
Similar articles
-
Changes in DNA methylation hallmark alterations in chromatin accessibility and gene expression for eye lens differentiation.Epigenetics Chromatin. 2022 Mar 5;15(1):8. doi: 10.1186/s13072-022-00440-z. Epigenetics Chromatin. 2022. PMID: 35246225 Free PMC article.
-
Multiomic analysis implicates FOXO4 in genetic regulation of chick lens fiber cell differentiation.Dev Biol. 2023 Dec;504:25-37. doi: 10.1016/j.ydbio.2023.09.005. Epub 2023 Sep 16. Dev Biol. 2023. PMID: 37722500 Free PMC article.
-
Profiling of chromatin accessibility and identification of general cis-regulatory mechanisms that control two ocular lens differentiation pathways.Epigenetics Chromatin. 2019 May 3;12(1):27. doi: 10.1186/s13072-019-0272-y. Epigenetics Chromatin. 2019. PMID: 31053165 Free PMC article.
-
Multiomics Analysis Reveals Novel Genetic Determinants for Lens Differentiation, Structure, and Transparency.Biomolecules. 2023 Apr 19;13(4):693. doi: 10.3390/biom13040693. Biomolecules. 2023. PMID: 37189439 Free PMC article. Review.
-
RNA-binding proteins and post-transcriptional regulation in lens biology and cataract: Mediating spatiotemporal expression of key factors that control the cell cycle, transcription, cytoskeleton and transparency.Exp Eye Res. 2022 Jan;214:108889. doi: 10.1016/j.exer.2021.108889. Epub 2021 Dec 11. Exp Eye Res. 2022. PMID: 34906599 Free PMC article. Review.
Cited by
-
A tamoxifen-inducible Cre knock-in mouse for lens-specific gene manipulation.Exp Eye Res. 2023 Jan;226:109306. doi: 10.1016/j.exer.2022.109306. Epub 2022 Nov 11. Exp Eye Res. 2023. PMID: 36372215 Free PMC article.
-
Lens regeneration in humans: using regenerative potential for tissue repairing.Ann Transl Med. 2020 Nov;8(22):1544. doi: 10.21037/atm-2019-rcs-03. Ann Transl Med. 2020. PMID: 33313289 Free PMC article. Review.
-
Ultrasound elastography for evaluating stiffness of the human lens nucleus with aging: a feasibility study.Int J Ophthalmol. 2021 Feb 18;14(2):240-244. doi: 10.18240/ijo.2021.02.09. eCollection 2021. Int J Ophthalmol. 2021. PMID: 33614452 Free PMC article.
-
Integrated single-cell multiomics uncovers foundational regulatory mechanisms of lens development and pathology.Development. 2024 Jan 1;151(1):dev202249. doi: 10.1242/dev.202249. Epub 2024 Jan 5. Development. 2024. PMID: 38180241 Free PMC article.
-
Lens Epithelial Explants Treated with Vitreous Humor Undergo Alterations in Chromatin Landscape with Concurrent Activation of Genes Associated with Fiber Cell Differentiation and Innate Immune Response.Cells. 2023 Feb 3;12(3):501. doi: 10.3390/cells12030501. Cells. 2023. PMID: 36766843 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

