ROR1 Potentiates FGFR Signaling in Basal-Like Breast Cancer
- PMID: 31137681
- PMCID: PMC6562526
- DOI: 10.3390/cancers11050718
ROR1 Potentiates FGFR Signaling in Basal-Like Breast Cancer
Erratum in
-
Correction: Pandey et al. ROR1 Potentiates FGFR Signaling in Basal-Like Breast Cancer. Cancers 2019, 11, 718.Cancers (Basel). 2022 Sep 19;14(18):4529. doi: 10.3390/cancers14184529. Cancers (Basel). 2022. PMID: 36139704 Free PMC article.
Abstract
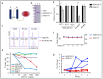
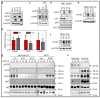
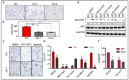
Among all breast cancer types, basal-like breast cancer (BLBC) represents an aggressive subtype that lacks targeted therapy. We and others have found that receptor tyrosine kinase-like orphan receptor 1 (ROR1) is overexpressed in BLBC and other types of cancer and that ROR1 is significantly correlated with patient prognosis. In addition, using primary patient-derived xenografts (PDXs) and ROR1-knockout BLBC cells, we found that ROR1+ cells form tumors in immunodeficient mice. We developed an anti-ROR1 immunotoxin and found that targeting ROR1 significantly kills ROR1+ cancer cells and slows down tumor growth in ROR1+ xenografts. Our bioinformatics analysis revealed that ROR1 expression is commonly associated with the activation of FGFR-mediated signaling pathway. Further biochemical analysis confirmed that ROR1 stabilized FGFR expression at the posttranslational level by preventing its degradation. CRISPR/Cas9-mediated ROR1 knockout significantly reduced cancer cell invasion at cellular levels by lowering FGFR protein and consequent inactivation of AKT. Our results identified a novel signaling regulation from ROR1 to FGFR and further confirm that ROR1 is a potential therapeutic target for ROR1+ BLBC cells.
Keywords: FGFR signaling; ROR1; breast cancer; cancer therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

