Engineering of Nanobodies Recognizing the Human Chemokine Receptor CCR7
- PMID: 31137829
- PMCID: PMC6566259
- DOI: 10.3390/ijms20102597
Engineering of Nanobodies Recognizing the Human Chemokine Receptor CCR7
Abstract
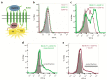
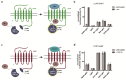
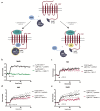
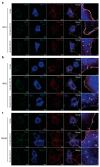
The chemokine receptor CCR7 plays a pivotal role in health and disease. In particular, CCR7 controls homing of antigen-bearing dendritic cells and T cells to lymph nodes, where adaptive immune responses are initiated. However, CCR7 also guides T cells to inflamed synovium and thereby contributes to rheumatoid arthritis and promotes cancer cell migration and metastasis formation. Nanobodies have recently emerged as versatile tools to study G-protein-coupled receptor functions and are being tested in diagnostics and therapeutics. In this study, we designed a strategy to engineer novel nanobodies recognizing human CCR7. We generated a nanobody library based on a solved crystal structure of the nanobody Nb80 recognizing the β2-adrenergic receptor (β2AR) and by specifically randomizing two segments within complementarity determining region 1 (CDR1) and CDR3 of Nb80 known to interact with β2AR. We fused the nanobody library to one half of split-YFP in order to identify individual nanobody clones interacting with CCR7 fused to the other half of split-YFP using bimolecular fluorescence complementation. We present three novel nanobodies, termed Nb1, Nb5, and Nb38, that recognize human CCR7 without interfering with G-protein-coupling and downstream signaling. Moreover, we were able to follow CCR7 trafficking upon CCL19 triggering using Nb1, Nb5, and Nb38.
Keywords: CCL19; CCR7; bimolecular fluorescence complementation; chemokine receptor; chemokines; nanobodies; split-luciferase complementation; β2-adrenergic receptor.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

