Genome-Wide Identification of Na+/H+ Antiporter (NHX) Genes in Sugar Beet (Beta vulgaris L.) and Their Regulated Expression under Salt Stress
- PMID: 31137880
- PMCID: PMC6562666
- DOI: 10.3390/genes10050401
Genome-Wide Identification of Na+/H+ Antiporter (NHX) Genes in Sugar Beet (Beta vulgaris L.) and Their Regulated Expression under Salt Stress
Abstract
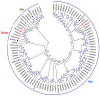
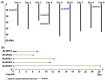
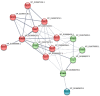
Salinity is one of the major environment factors that limits the growth of plants and the productivity of crops worldwide. It has been shown that Na+ transporters play a central role in salt tolerance and development of plants. The objective of this study was to identify Na+/H+ antiporter (NHX) genes and investigate their expression patterns in sugar beet (Beta vulgaris L.) subjected to various concentrations of NaCl. A total of five putative NHX genes were identified and distributed on four chromosomes in sugar beet. Phylogenetic analysis revealed that these BvNHX genes are grouped into three major classes, viz Vac- (BvNHX1, -2 and -3), Endo- (BvNHX4), and PM-class NHX (BvNHX5/BvSOS1), and within each class the exon/intron structures are conserved. The amiloride-binding site is found in TM3 at N-terminus of Vac-class NHX proteins. Protein-protein interaction (PPI) prediction suggested that only BvNHX5 putatively interacts with calcineurin B-like proteins (CBL) and CBL-interacting protein kinases (CIPK), implying it might be the primary NHX involved in CBL-CIPK pathway under saline condition. It was also found that BvNHX5 contains one abscisic acid (ABA)-responsive element (ABRE), suggesting that BvNHX5 might be involved in ABA signal responsiveness. Additionally, the qRT-PCR analysis showed that all the BvNHX genes in both roots and leaves are significantly up-regulated by salt, and the transcription levels under high salinity are significantly higher than those under either low or moderate salinity. Taken together, this work gives a detailed overview of the BvNHX genes and their expression patterns under salt stress. Our findings also provide useful information for elucidating the molecular mechanisms of Na+ homeostasis and further functional identification of the BvNHX genes in sugar beet.
Keywords: Na+ compartmentalization; Na+ exclusion; Na+/H+ antiporter; amiloride-binding site; salt tolerance; sugar beet.
Conflict of interest statement
The authors declare that they have no conflict of interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

