Complex Gene Regulation Underlying Mineral Nutrient Homeostasis in Soybean Root Response to Acidity Stress
- PMID: 31137896
- PMCID: PMC6563148
- DOI: 10.3390/genes10050402
Complex Gene Regulation Underlying Mineral Nutrient Homeostasis in Soybean Root Response to Acidity Stress
Abstract
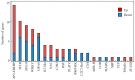
Proton toxicity is one of the major environmental stresses limiting crop production and becomes increasingly serious because of anthropogenic activities. To understand acid tolerance mechanisms, the plant growth, mineral nutrients accumulation, and global transcriptome changes in soybean (Glycine max) in response to long-term acidity stress were investigated. Results showed that acidity stress significantly inhibited soybean root growth but exhibited slight effects on the shoot growth. Moreover, concentrations of essential mineral nutrients were significantly affected by acidity stress, mainly differing among soybean organs and mineral nutrient types. Concentrations of phosphorus (P) and molybdenum (Mo) in both leaves and roots, nitrogen (N), and potassium (K) in roots and magnesium (Mg) in leaves were significantly decreased by acidity stress, respectively. Whereas, concentrations of calcium (Ca), sulfate (S), and iron (Fe) were increased in both leaves and roots. Transcriptome analyses in soybean roots resulted in identification of 419 up-regulated and 555 down-regulated genes under acid conditions. A total of 38 differentially expressed genes (DEGs) were involved in mineral nutrients transportation. Among them, all the detected five GmPTs, four GmZIPs, two GmAMTs, and GmKUPs, together with GmIRT1, GmNramp5, GmVIT2.1, GmSKOR, GmTPK5, and GmHKT1, were significantly down-regulated by acidity stress. Moreover, the transcription of genes encoding transcription factors (e.g., GmSTOP2s) and associated with pH stat metabolic pathways was significantly up-regulated by acidity stress. Taken together, it strongly suggests that maintaining pH stat and mineral nutrient homeostasis are adaptive strategies of soybean responses to acidity stress, which might be regulated by a complex signaling network.
Keywords: acidity stress; mineral nutrient; roots; soybean; transporter.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Von Uexküll H.R., Mutert E. Global extent, development and economic impact of acid soils. Plant Soil. 1995;171:1–15. doi: 10.1007/BF00009558. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

