Clostridium difficile ribotype 017 - characterization, evolution and epidemiology of the dominant strain in Asia
- PMID: 31138041
- PMCID: PMC6542179
- DOI: 10.1080/22221751.2019.1621670
Clostridium difficile ribotype 017 - characterization, evolution and epidemiology of the dominant strain in Asia
Abstract
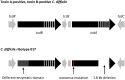
Clostridium difficile ribotype (RT) 017 is an important toxigenic C. difficile RT which, due to a deletion in the repetitive region of the tcdA gene, only produces functional toxin B. Strains belonging to this RT were initially dismissed as nonpathogenic and circulated largely undetected for almost two decades until they rose to prominence following a series of outbreaks in the early 2000s. Despite lacking a functional toxin A, C. difficile RT 017 strains have been shown subsequently to be capable of causing disease as severe as that caused by strains producing both toxins A and B. While C. difficile RT 017 strains can be found in almost every continent today, epidemiological studies suggest that the RT is endemic in Asia and that the global spread of this MLST clade 4 lineage member is a relatively recent event. C. difficile RT 017 transmission appears to be mostly from human to human with only a handful of reports of isolations from animals. An important feature of C. difficile RT 017 strains is their resistance to several antimicrobials and this has been documented as a possible factor driving multiple outbreaks in different parts of the world. This review summarizes what is currently known regarding the emergence and evolution of strains belonging to C. difficile RT 017 as well as features that have allowed it to become an RT of global importance.
Keywords: epidemiology; ribotype 017.
Figures



Similar articles
-
High prevalence and diversity of tcdA-negative and tcdB-positive, and non-toxigenic, Clostridium difficile in Thailand.Anaerobe. 2019 Jun;57:4-10. doi: 10.1016/j.anaerobe.2019.03.008. Epub 2019 Mar 9. Anaerobe. 2019. PMID: 30862468
-
Phenotypic characterisation of Clostridium difficile PCR ribotype 251, an emerging multi-locus sequence type clade 2 strain in Australia.Anaerobe. 2019 Dec;60:102066. doi: 10.1016/j.anaerobe.2019.06.019. Epub 2019 Jun 28. Anaerobe. 2019. PMID: 31260740
-
A series of three cases of severe Clostridium difficile infection in Australia associated with a binary toxin producing clade 2 ribotype 251 strain.Anaerobe. 2019 Feb;55:117-123. doi: 10.1016/j.anaerobe.2018.11.009. Epub 2018 Nov 27. Anaerobe. 2019. PMID: 30500477
-
Reprint of New opportunities for improved ribotyping of C. difficile clinical isolates by exploring their genomes.J Microbiol Methods. 2013 Dec;95(3):425-40. doi: 10.1016/j.mimet.2013.09.009. Epub 2013 Sep 16. J Microbiol Methods. 2013. PMID: 24050948 Review.
-
New opportunities for improved ribotyping of C. difficile clinical isolates by exploring their genomes.J Microbiol Methods. 2013 Jun;93(3):257-72. doi: 10.1016/j.mimet.2013.02.013. Epub 2013 Mar 29. J Microbiol Methods. 2013. PMID: 23545446 Review.
Cited by
-
Clostridioides difficile Sporulation.Adv Exp Med Biol. 2024;1435:273-314. doi: 10.1007/978-3-031-42108-2_13. Adv Exp Med Biol. 2024. PMID: 38175480 Review.
-
Global evolutionary dynamics and resistome analysis of Clostridioides difficile ribotype 017.Microb Genom. 2022 Mar;8(3):000792. doi: 10.1099/mgen.0.000792. Microb Genom. 2022. PMID: 35316173 Free PMC article.
-
Clostridioides difficile infection epidemiology during the COVID-19 pandemic in Greece.Future Microbiol. 2024 Sep;19(13):1119-1127. doi: 10.1080/17460913.2024.2358653. Epub 2024 Jun 24. Future Microbiol. 2024. PMID: 38913938 Free PMC article.
-
Make It Less difficile: Understanding Genetic Evolution and Global Spread of Clostridioides difficile.Genes (Basel). 2022 Nov 24;13(12):2200. doi: 10.3390/genes13122200. Genes (Basel). 2022. PMID: 36553467 Free PMC article. Review.
-
An epidemiological surveillance study (2021-2022): detection of a high diversity of Clostridioides difficile isolates in one tertiary hospital in Chongqing, Southwest China.BMC Infect Dis. 2023 Oct 19;23(1):703. doi: 10.1186/s12879-023-08666-2. BMC Infect Dis. 2023. PMID: 37858038 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
