The FANCM-BLM-TOP3A-RMI complex suppresses alternative lengthening of telomeres (ALT)
- PMID: 31138797
- PMCID: PMC6538672
- DOI: 10.1038/s41467-019-10180-6
The FANCM-BLM-TOP3A-RMI complex suppresses alternative lengthening of telomeres (ALT)
Erratum in
-
Author Correction: The FANCM-BLM-TOP3A-RMI complex suppresses alternative lengthening of telomeres (ALT).Nat Commun. 2019 Nov 20;10(1):5345. doi: 10.1038/s41467-019-13097-2. Nat Commun. 2019. PMID: 31745078 Free PMC article.
Abstract
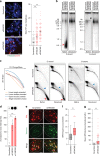
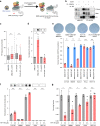
The collapse of stalled replication forks is a major driver of genomic instability. Several committed mechanisms exist to resolve replication stress. These pathways are particularly pertinent at telomeres. Cancer cells that use Alternative Lengthening of Telomeres (ALT) display heightened levels of telomere-specific replication stress, and co-opt stalled replication forks as substrates for break-induced telomere synthesis. FANCM is a DNA translocase that can form independent functional interactions with the BLM-TOP3A-RMI (BTR) complex and the Fanconi anemia (FA) core complex. Here, we demonstrate that FANCM depletion provokes ALT activity, evident by increased break-induced telomere synthesis, and the induction of ALT biomarkers. FANCM-mediated attenuation of ALT requires its inherent DNA translocase activity and interaction with the BTR complex, but does not require the FA core complex, indicative of FANCM functioning to restrain excessive ALT activity by ameliorating replication stress at telomeres. Synthetic inhibition of FANCM-BTR complex formation is selectively toxic to ALT cancer cells.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. - PubMed
-
- Schmutz I, Timashev L, Xie W, Patel DJ, de Lange T. TRF2 binds branched DNA to safeguard telomere integrity. Nat. Struct. Mol. Biol. 2017;24:734–742. - PubMed
-
- Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

