Neutrophil Extracellular Traps protein composition is specific for patients with Lupus nephritis and includes methyl-oxidized αenolase (methionine sulfoxide 93)
- PMID: 31138830
- PMCID: PMC6538718
- DOI: 10.1038/s41598-019-44379-w
Neutrophil Extracellular Traps protein composition is specific for patients with Lupus nephritis and includes methyl-oxidized αenolase (methionine sulfoxide 93)
Abstract
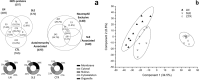
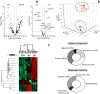
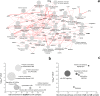
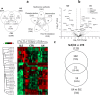
NETs constitute a network of DNA and proteins released by neutrophils in response to infectious and immunologic triggers. NET proteins are recognized as autoantigens in ANCA vasculitis; limited knowledge is available in other autoimmune pathologies. The composition of NETs produced ex vivo by resting and Phorbol-myristate acetate (PMA) stimulated neutrophils was analyzed by high-throughput Fusion Orbitrap technology in 16 patients with Systemic Lupus Erythematosus/Lupus nephritis (9 SLE/7 LN) and in 11 controls. Seven-hundred proteins were characterized and specific fingerprints discriminated LN from SLE. We focused on methyl-oxidized αenolase (methionine sulfoxide 93) that was markedly increased in NETs from LN and was localized in NET filaments in tight connection and outlying DNA. The isotype of anti-αenolase antibodies was IgG2 in LN and IgG4 in other autoimmune glomerulonephritis (Membranous Nephropathy, MN); serum anti-αenolase IgG2 were higher in LN than in SLE and absent in MN. The same IgG2 antibodies recognized 5 epitopes of the protein one containing methionine sulphoxide 93. In conclusion, specific NET protein fingerprints characterize different subsets of SLE; methyl-oxidized αenolase is over-expressed in LN. Circulating anti-αenolase IgG2 recognize the oxidized epitope and are high in serum of LN patients. Post-translational modified NET proteins contribute to autoimmunity in patients with LN.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

