Immune Correlates of Disease Progression in Linked HIV-1 Infection
- PMID: 31139189
- PMCID: PMC6527802
- DOI: 10.3389/fimmu.2019.01062
Immune Correlates of Disease Progression in Linked HIV-1 Infection
Abstract
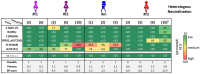
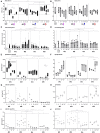
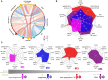
Genetic and immunologic analyses of epidemiologically-linked HIV transmission enable insights into the impact of immune responses on clinical outcomes. Human vaccine trials and animal studies of HIV-1 infection have suggested immune correlates of protection; however, their role in natural infection in terms of protection from disease progression is mostly unknown. Four HIV-1+ Cameroonian individuals, three of them epidemiologically-linked in a polygamous heterosexual relationship and one incidence-matched case, were studied over 15 years for heterologous and cross-neutralizing antibody responses, antibody binding, IgA/IgG levels, antibody-dependent cellular cytotoxicity (ADCC) against cells expressing wild-type or CD4-bound Env, viral evolution, Env epitopes, and host factors including HLA-I alleles. Despite viral infection with related strains, the members of the transmission cluster experienced contrasting clinical outcomes including cases of rapid progression and long-term non-progression in the absence of strongly protective HLA-I or CCR5Δ32 alleles. Slower progression and higher CD4/CD8 ratios were associated with enhanced IgG antibody binding to native Env and stronger V1V2 antibody binding responses in the presence of viruses with residue K169 in V2. ADCC against cells expressing Env in the CD4-bound conformation in combination with low Env-specific IgA/IgG ratios correlated with better clinical outcome. This data set highlights for the first time that V1V2-directed antibody responses and ADCC against cells expressing open, CD4-exposed Env, in the presence of low plasma IgA/IgG ratios, can correlate with clinical outcome in natural infection. These parameters are comparable to the major correlates of protection, identified post-hoc in the RV144 vaccine trial; thus, they may also modulate the rate of clinical progression once infected. The findings illustrate the potential of immune correlate analysis in natural infection to guide vaccine development.
Keywords: ADCC; BEAST; IgA/IgG ratio; V1V2 antibody binding; epidemiologically-linked infection; human immunodeficiency virus (HIV); protective immune parameters and host factors; viral signature K169.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

