Defining molecular risk in ALK+ NSCLC
- PMID: 31139322
- PMCID: PMC6517105
- DOI: 10.18632/oncotarget.26886
Defining molecular risk in ALK+ NSCLC
Abstract
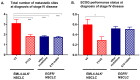
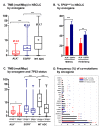
Anaplastic lymphoma kinase (ALK)-positive non-small-cell lung cancers (NSCLC) have the best prognosis among metastatic pulmonary malignancies, with a median patient survival currently exceeding 5 years. While this is definitely a major therapeutic success for thoracic oncology, it may not be entirely attributable to rapid drug development and the strenuous clinical efforts. At the genetic level, ALK+ disease is also unique, distinguished by the lowest tumor mutational burden (mean below 3 mutations/Mbp), the lowest frequency of TP53 mutations (20-25%) and very few other co-mutations compared to other NSCLC. The relative simplicity and stability of the genetic landscape not only contribute to the relatively favourable clinical course, but also make study of the effects from individual molecular features easier. EML4-ALK fusion variant 3 (E6;A20) and TP53 mutations were recently identified as main molecular determinants of adverse outcome: they occur in about 30-40% and 20-25% of newly-diagnosed cases, respectively, have possibly synergistic effects and are independently associated with more aggressive disease, shorter progression-free survival under treatment with ALK inhibitors and worse overall survival. Secondary detection of TP53 mutations at disease progression in previously negative patients defines another subset (about 20%) with similarly poor outcome, while detection of ALK resistance mutations guides next-line therapy. As our biological understanding deepens, additional molecular risk factors will be identified and refine our concepts further. The translation of clinical risk at the molecular level and the ability to predict early events are of key importance for individualized patient management and preclinical modeling in order to advance therapeutic options.
Keywords: ALK+ non-small cell lung cancer; EML4-ALK fusion variant; TP53 mutation; overall survival; treatment resistance.
Conflict of interest statement
CONFLICTS OF INTEREST PC reports lecture fees from Roche, Chugai and Novartis; SD reports lecture fees from Roche; HS reports advisory board and speaker’s honoraria from Roche; MT reports advisory board honoraria from Novartis, Lilly, BMS, MSD, Roche, Celgene, Takeda, AbbVie, Boehringer, speaker’s honoraria from Lilly, MSD, Takeda, research funding from Ast raZeneca, BMS, Celgene, Novartis, Roche and travel grants from BMS, MSD, Novartis, Boehringer; AS reports advisory board honoraria from Bayer, Novartis, AstraZeneca, ThermoFisher, BMS, Illumina, and lecture fees from Bayer, Roche, BMS, Illumina, AstraZeneca, Novartis, ThermoFisher; MK and JB declare no potential conflicts of interest.
Figures

References
-
- Duruisseaux M, Besse B, Cadranel J, Perol M, Mennecier B, Bigay-Game L, Descourt R, Dansin E, Audigier-Valette C, Moreau L, Hureaux J, Veillon R, Otto J, et al. Overall survival with crizotinib and next-generation ALK inhibitors in ALK-positive non-small-cell lung cancer (IFCT-1302 CLINALK): a French nationwide cohort retrospective study. Oncotarget. 2017;8:21903–17. doi: 10.18632/oncotarget.15746. - DOI - PMC - PubMed
-
- Kayaniyil S, Hurry M, Wilson J, Wheatley-Price P, Melosky B, Rothenstein J, Cohen V, Koch C, Zhang J, Osenenko K, Liu G. Treatment patterns and survival in patients with ALK-positive non-small-cell lung cancer: a Canadian retrospective study. Curr Oncol. 2016;23:e589–e97. doi: 10.3747/co.23.3273. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

