Pairwise visual comparison of small RNA secondary structures with base pair probabilities
- PMID: 31142266
- PMCID: PMC6542128
- DOI: 10.1186/s12859-019-2902-6
Pairwise visual comparison of small RNA secondary structures with base pair probabilities
Abstract
Background: Predicted RNA secondary structures are typically visualized using dot-plots for base pair binding probabilities and planar graphs for unique structures, such as the minimum free energy structure. These are however difficult to analyze simultaneously.
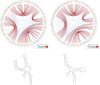
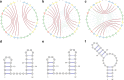
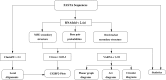
Results: This work introduces a compact unified view of the most stable conformation of an RNA secondary structure and its base pair probabilities, which is called the Circular Secondary Structure Base Pairs Probabilities Plot (CS2BP2-Plot). Along with our design we provide access to a web server implementation of our solution that facilitates pairwise comparison of short RNA (and DNA) sequences up to 200 base pairs. The web server first calculates the minimum free energy secondary structure and the base pair probabilities for up to 10 RNA or DNA sequences using RNAfold and then provides a two panel comparative view that includes CS2BP2-Plots along with the traditional graph, planar and circular diagrams obtained with VARNA. The CS2BP2-Plots include highlighting of the nucleotide differences between two selected sequences using ClustalW local alignments. We also provide descriptive statistics, dot-bracket secondary structure representations and ClustalW local alignments for compared sequences.
Conclusions: Using circular diagrams and colour and weight-coded arcs, we demonstrate how a single image can replace the state-of-the-art dual representations (dot-plots and minimum free energy structures) for base-pair probabilities of RNA secondary structures while allowing efficient exploration and comparison of different RNA conformations via a web server front end. With that, we provide the community, especially the biologically oriented, with an intuitive tool for ncRNA visualization. Web-server: https://nrcmonsrv01.nrc.ca/cs2bp2plot.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

