Atypical function of a centrosomal module in WNT signalling drives contextual cancer cell motility
- PMID: 31142743
- PMCID: PMC6541620
- DOI: 10.1038/s41467-019-10241-w
Atypical function of a centrosomal module in WNT signalling drives contextual cancer cell motility
Erratum in
-
Author Correction: Atypical function of a centrosomal module in WNT signalling drives contextual cancer cell motility.Nat Commun. 2025 Feb 26;16(1):1981. doi: 10.1038/s41467-025-57127-8. Nat Commun. 2025. PMID: 40011433 Free PMC article. No abstract available.
Abstract
Centrosomes control cell motility, polarity and migration that is thought to be mediated by their microtubule-organizing capacity. Here we demonstrate that WNT signalling drives a distinct form of non-directional cell motility that requires a key centrosome module, but not microtubules or centrosomes. Upon exosome mobilization of PCP-proteins, we show that DVL2 orchestrates recruitment of a CEP192-PLK4/AURKB complex to the cell cortex where PLK4/AURKB act redundantly to drive protrusive activity and cell motility. This is mediated by coordination of formin-dependent actin remodelling through displacement of cortically localized DAAM1 for DAAM2. Furthermore, abnormal expression of PLK4, AURKB and DAAM1 is associated with poor outcomes in breast and bladder cancers. Thus, a centrosomal module plays an atypical function in WNT signalling and actin nucleation that is critical for cancer cell motility and is associated with more aggressive cancers. These studies have broad implications in how contextual signalling controls distinct modes of cell migration.
Conflict of interest statement
The authors declare no competing interests.
Figures
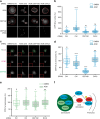
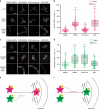
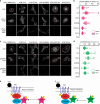
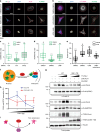
Similar articles
-
DAAM1 is a formin required for centrosome re-orientation during cell migration.PLoS One. 2010 Sep 29;5(9):e13064. doi: 10.1371/journal.pone.0013064. PLoS One. 2010. PMID: 20927366 Free PMC article.
-
Cep192 controls the balance of centrosome and non-centrosomal microtubules during interphase.PLoS One. 2014 Jun 27;9(6):e101001. doi: 10.1371/journal.pone.0101001. eCollection 2014. PLoS One. 2014. PMID: 24971877 Free PMC article.
-
Planar cell polarity pathway regulates actin rearrangement, cell shape, motility, and nephrin distribution in podocytes.Am J Physiol Renal Physiol. 2011 Feb;300(2):F549-60. doi: 10.1152/ajprenal.00566.2009. Epub 2010 Jun 9. Am J Physiol Renal Physiol. 2011. PMID: 20534871
-
TRIM37: a critical orchestrator of centrosome function.Cell Cycle. 2021 Dec;20(23):2443-2451. doi: 10.1080/15384101.2021.1988289. Epub 2021 Oct 21. Cell Cycle. 2021. PMID: 34672905 Free PMC article. Review.
-
Dishevelled: A masterful conductor of complex Wnt signals.Cell Signal. 2018 Jul;47:52-64. doi: 10.1016/j.cellsig.2018.03.004. Epub 2018 Mar 17. Cell Signal. 2018. PMID: 29559363 Free PMC article. Review.
Cited by
-
Modelling the Functions of Polo-Like Kinases in Mice and Their Applications as Cancer Targets with a Special Focus on Ovarian Cancer.Cells. 2021 May 12;10(5):1176. doi: 10.3390/cells10051176. Cells. 2021. PMID: 34065956 Free PMC article. Review.
-
Exosomes: new targets for understanding axon guidance in the developing central nervous system.Front Cell Dev Biol. 2025 Jan 9;12:1510862. doi: 10.3389/fcell.2024.1510862. eCollection 2024. Front Cell Dev Biol. 2025. PMID: 39850798 Free PMC article. Review.
-
Centriole Number and the Accumulation of Microtubules Modulate the Timing of Apical Insertion during Radial Intercalation.Curr Biol. 2020 May 18;30(10):1958-1964.e3. doi: 10.1016/j.cub.2020.03.013. Epub 2020 Apr 2. Curr Biol. 2020. PMID: 32243862 Free PMC article.
-
Independent prognostic value of CLDN6 in bladder cancer based on M2 macrophages related signature.iScience. 2024 Feb 5;27(3):109138. doi: 10.1016/j.isci.2024.109138. eCollection 2024 Mar 15. iScience. 2024. PMID: 38380255 Free PMC article.
-
Lipid-associated macrophages in the tumor-adipose microenvironment facilitate breast cancer progression.Oncoimmunology. 2022 Jun 8;11(1):2085432. doi: 10.1080/2162402X.2022.2085432. eCollection 2022. Oncoimmunology. 2022. PMID: 35712121 Free PMC article.
References
-
- Friedl, P. & Alexander, S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell147, 992–1009 (2011). - PubMed
-
- Yilmaz, M. & Christofori, G. Mechanisms of motility in metastasizing cells. Mol. Cancer Res8, 629–642 (2010). - PubMed
-
- Ridley, A. Life at the leading edge. Cell145, 1012–1022 (2011). - PubMed
-
- Goode, B. & Eck, M. Mechanism and function of formins in the control of actin assembly. Annu Rev. Biochem76, 593–627 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- FDN143252/Gouvernement du Canada | Canadian Institutes of Health Research (Instituts de Recherche en Santé du Canada)/International
- 704378/Canadian Cancer Society Research Institute (Société Canadienne du Cancer)/International
- 702320/Canadian Cancer Society Research Institute (Société Canadienne du Cancer)/International
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

