The vaginal microbiome and preterm birth
- PMID: 31142849
- PMCID: PMC6750801
- DOI: 10.1038/s41591-019-0450-2
The vaginal microbiome and preterm birth
Abstract
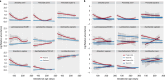
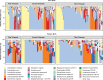
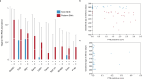
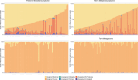
The incidence of preterm birth exceeds 10% worldwide. There are significant disparities in the frequency of preterm birth among populations within countries, and women of African ancestry disproportionately bear the burden of risk in the United States. In the present study, we report a community resource that includes 'omics' data from approximately 12,000 samples as part of the integrative Human Microbiome Project. Longitudinal analyses of 16S ribosomal RNA, metagenomic, metatranscriptomic and cytokine profiles from 45 preterm and 90 term birth controls identified harbingers of preterm birth in this cohort of women predominantly of African ancestry. Women who delivered preterm exhibited significantly lower vaginal levels of Lactobacillus crispatus and higher levels of BVAB1, Sneathia amnii, TM7-H1, a group of Prevotella species and nine additional taxa. The first representative genomes of BVAB1 and TM7-H1 are described. Preterm-birth-associated taxa were correlated with proinflammatory cytokines in vaginal fluid. These findings highlight new opportunities for assessment of the risk of preterm birth.
Conflict of interest statement
J.K. and Y.C.T. are full-time employees at Pacific Biosciences, a company developing single-molecule sequencing technologies. No other authors report any competing interests.
Figures









Comment in
-
Gestational shaping of the maternal vaginal microbiome.Nat Med. 2019 Jun;25(6):882-883. doi: 10.1038/s41591-019-0483-6. Nat Med. 2019. PMID: 31142848 No abstract available.
-
Tracking humans and microbes.Nature. 2019 May;569(7758):632-633. doi: 10.1038/d41586-019-01591-y. Nature. 2019. PMID: 31142867 No abstract available.
-
After the Integrative Human Microbiome Project, what's next for the microbiome community?Nature. 2019 May;569(7758):599. doi: 10.1038/d41586-019-01674-w. Nature. 2019. PMID: 31142868 No abstract available.
References
-
- Behrman, R. E. & Butler, A. S. (eds), for Institute of Medicine Committee on Understanding Premature Birth and Assuring Healthy Outcomes. Societal Costs of Preterm Birth (National Academies Press, 2007).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

