Phenotyping to Facilitate Accrual for a Cardiovascular Intervention
- PMID: 31143314
- PMCID: PMC6522233
- DOI: 10.14740/jocmr3830
Phenotyping to Facilitate Accrual for a Cardiovascular Intervention
Abstract
Background: The conventional approach for clinical studies is to identify a cohort of potentially eligible patients and then screen for enrollment. In an effort to reduce the cost and manual effort involved in the screening process, several studies have leveraged electronic health records (EHR) to refine cohorts to better match the eligibility criteria, which is referred to as phenotyping. We extend this approach to dynamically identify a cohort by repeating phenotyping in alternation with manual screening.
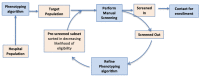
Methods: Our approach consists of multiple screen cycles. At the start of each cycle, the phenotyping algorithm is used to identify eligible patients from the EHR, creating an ordered list such that patients that are most likely eligible are listed first. This list is then manually screened, and the results are analyzed to improve the phenotyping for the next cycle. We describe the preliminary results and challenges in the implementation of this approach for an intervention study on heart failure.
Results: A total of 1,022 patients were screened, with 223 (23%) of patients being found eligible for enrollment into the intervention study. The iterative approach improved the phenotyping in each screening cycle. Without an iterative approach, the positive screening rate (PSR) was expected to dip below the 20% measured in the first cycle; however, the cyclical approach increased the PSR to 23%.
Conclusions: Our study demonstrates that dynamic phenotyping can facilitate recruitment for prospective clinical study. Future directions include improved informatics infrastructure and governance policies to enable real-time updates to research repositories, tooling for EHR annotation, and methodologies to reduce human annotation.
Keywords: Cohort identification; Electronic health records; Intervention; Phenotyping.
Conflict of interest statement
The authors have no conflict of interest to disclose.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
