A Central Region of NF-κB Essential Modulator Is Required for IKKβ-Induced Conformational Change and for Signal Propagation
- PMID: 31145594
- PMCID: PMC9295417
- DOI: 10.1021/acs.biochem.8b01316
A Central Region of NF-κB Essential Modulator Is Required for IKKβ-Induced Conformational Change and for Signal Propagation
Abstract
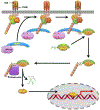
NF-κB essential modulator (NEMO) regulates NF-κB signaling by acting as a scaffold for the kinase IKKβ to direct its activity toward the NF-κB inhibitor, IκBα. Here, we show that a highly conserved central region of NEMO termed the intervening domain (IVD, amino acids 112-195) plays a key role in NEMO function. We determined a structural model of full-length NEMO by small-angle X-ray scattering and show that full-length, wild-type NEMO becomes more compact upon binding of a peptide comprising the NEMO binding domain of IKKβ (amino acids 701-745). Mutation of conserved IVD residues (9SG-NEMO) disrupts this conformational change in NEMO and abolishes the ability of NEMO to propagate NF-κB signaling in cells, although the affinity of 9SG-NEMO for IKKβ compared to that of the wild type is unchanged. On the basis of these results, we propose a model in which the IVD is required for a conformational change in NEMO that is necessary for its ability to direct phosphorylation of IκBα by IKKβ. Our findings suggest a molecular explanation for certain disease-associated mutations within the IVD and provide insight into the role of conformational change in signaling scaffold proteins.
Figures



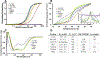



References
-
- Shaw AS, and Filbert EL (2009) Scaffold proteins and immune-cell signalling. Nat Rev Immunol 9, 47–56. - PubMed
-
- Pan CQ, Sudol M, Sheetz M, and Low BC (2012) Modularity and functional plasticity of scaffold proteins as p(l)acemakers in cell signaling. Cell Signal 24, 2143–2165. - PubMed
-
- Dhanasekaran DN, Kashef K, Lee CM, Xu H, and Reddy EP (2007) Scaffold proteins of MAP-kinase modules. Oncogene 26, 3185–3202. - PubMed
-
- Vaquero J, Nguyen Ho-Bouldoires TH, Clapéron A, and Fouassier L (2017) Role of the PDZ-scaffold protein NHERF1/EBP50 in cancer biology: from signaling regulation to clinical relevance. Oncogene 3067–3079. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

