Parameter estimation and identifiability in a neural population model for electro-cortical activity
- PMID: 31145724
- PMCID: PMC6542506
- DOI: 10.1371/journal.pcbi.1006694
Parameter estimation and identifiability in a neural population model for electro-cortical activity
Abstract
Electroencephalography (EEG) provides a non-invasive measure of brain electrical activity. Neural population models, where large numbers of interacting neurons are considered collectively as a macroscopic system, have long been used to understand features in EEG signals. By tuning dozens of input parameters describing the excitatory and inhibitory neuron populations, these models can reproduce prominent features of the EEG such as the alpha-rhythm. However, the inverse problem, of directly estimating the parameters from fits to EEG data, remains unsolved. Solving this multi-parameter non-linear fitting problem will potentially provide a real-time method for characterizing average neuronal properties in human subjects. Here we perform unbiased fits of a 22-parameter neural population model to EEG data from 82 individuals, using both particle swarm optimization and Markov chain Monte Carlo sampling. We estimate how much is learned about individual parameters by computing Kullback-Leibler divergences between posterior and prior distributions for each parameter. Results indicate that only a single parameter, that determining the dynamics of inhibitory synaptic activity, is directly identifiable, while other parameters have large, though correlated, uncertainties. We show that the eigenvalues of the Fisher information matrix are roughly uniformly spaced over a log scale, indicating that the model is sloppy, like many of the regulatory network models in systems biology. These eigenvalues indicate that the system can be modeled with a low effective dimensionality, with inhibitory synaptic activity being prominent in driving system behavior.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





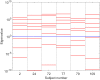
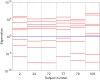



References
-
- Kropotov JD. Chapter 2—Alpha Rhythms In: Kropotov JD, editor. Quantitative EEG, Event-Related Potentials and Neurotherapy. San Diego: Academic Press; 2009. p. 29–58. Available from: http://www.sciencedirect.com/science/article/pii/B9780123745125000025.
-
- Aminoff MJ. Chapter 3—Electroencephalography: General Principles and Clinical Applications In: Aminoff MJ, editor. Aminoff’s Electrodiagnosis in Clinical Neurology (Sixth Edition) sixth edition ed. London: W.B. Saunders; 2012. p. 37–84. Available from: http://www.sciencedirect.com/science/article/pii/B9781455703081000030.
-
- Berger H. Über das elektrenkephalogramm des menschen. Archiv für psychiatrie und nervenkrankheiten. 1929;87(1):527–570. 10.1007/BF01797193 - DOI
-
- Berger H. On the electroencephalogram of man. Third Report 1931; Twelfth Report 1937. Translated by Pierre Gloor. Electroencephalogr Clin Neurophysiol. 1931;28(suppl):113–167.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

