The Emergence and Decennary Distribution of Clade 2.3.4.4 HPAI H5Nx
- PMID: 31146461
- PMCID: PMC6616411
- DOI: 10.3390/microorganisms7060156
The Emergence and Decennary Distribution of Clade 2.3.4.4 HPAI H5Nx
Abstract
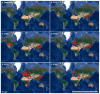
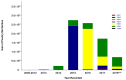
Reassortment events among influenza viruses occur naturally and may lead to the development of new and different subtypes which often ignite the possibility of an influenza outbreak. Between 2008 and 2010, highly pathogenic avian influenza (HPAI) H5 of the N1 subtype from the A/goose/Guangdong/1/96-like (Gs/GD) lineage generated novel reassortants by introducing other neuraminidase (NA) subtypes reported to cause most outbreaks in poultry. With the extensive divergence of the H5 hemagglutinin (HA) sequences of documented viruses, the WHO/FAO/OIE H5 Evolutionary Working Group clustered these viruses into a systematic and unified nomenclature of clade 2.3.4.4 currently known as "H5Nx" viruses. The rapid emergence and circulation of these viruses, namely, H5N2, H5N3, H5N5, H5N6, H5N8, and the regenerated H5N1, are of great concern based on their pandemic potential. Knowing the evolution and emergence of these novel reassortants helps to better understand their complex nature. The eruption of reports of each H5Nx reassortant through time demonstrates that it could persist beyond its usual seasonal activity, intensifying the possibility of these emerging viruses' pandemic potential. This review paper provides an overview of the emergence of each novel HPAI H5Nx virus as well as its current epidemiological distribution.
Keywords: H5Nx; avian; avian influenza; dissemination; epidemiology; evolution.
Conflict of interest statement
The authors declare no conflict of interests.
Figures

References
-
- Dhingra M.S., Artois J., Robinson T.P., Linard C., Chaiban C., Xenarios I., Engler R., Liechti R., Kuznetsov D., Xiao X., et al. Global mapping of highly pathogenic avian influenza H5N1 and H5Nx clade 2.3.4.4 viruses with spatial cross-validation. eLife. 2016;5 doi: 10.7554/eLife.19571. - DOI - PMC - PubMed
-
- Duan L., Campitelli L., Fan X.H., Leung Y.H., Vijaykrishna D., Zhang J.X., Donatelli I., Delogu M., Li K.S., Foni E., et al. Characterization of low-pathogenic H5 subtype influenza viruses from Eurasia: Implications for the origin of highly pathogenic H5N1 viruses. J. Virol. 2007;81:7529–7539. doi: 10.1128/JVI.00327-07. - DOI - PMC - PubMed
-
- Hill N.J., Hussein I.T., Davis K.R., Ma E.J., Spivey T.J., Ramey A.M., Puryear W.B., Das S.R., Halpin R.A., Lin X., et al. Reassortment of Influenza A Viruses in Wild Birds in Alaska before H5 Clade 2.3.4.4 Outbreaks. Emerg. Infect. Dis. 2017;23:654–657. doi: 10.3201/eid2304.161668. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

