Comparative transcriptomic analysis reveals genetic divergence and domestication genes in Diospyros
- PMID: 31146695
- PMCID: PMC6543618
- DOI: 10.1186/s12870-019-1839-2
Comparative transcriptomic analysis reveals genetic divergence and domestication genes in Diospyros
Abstract
Background: Persimmon (Diospyros kaki) is the most economically cultivated species belonging to the genus Diospyros. However, little is known about the interspecific diversity and mechanism of domestication, partly due to the lack of genomic information that is available for closely related species of D. kaki (DK). Here, we performed transcriptome sequencing on nine samples, including DK, a variety of DK and seven closely related species, to evaluate the interspecific genetic divergence and to identify candidate genes involved in persimmon domestication.
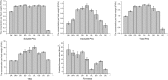
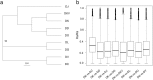
Results: We obtained a total of 483,421 unigenes with N50 at 1490 bp in the nine Diospyros samples and identified 2603 orthogroups that were shared among all the samples using OrthoMCL analysis. A phylogenetic tree was established based on the tandem 2603 one-to-one single copy gene alignments, showing that DK was closely related to D. kaki var. silvestris (DKV) and that it clustered with the clade of D. deyangnsis (DD) and was farthest from the D. cathayensis (DC) species. The nonsynonymous substitutions (Ka), via synonymous substitution (Ks) ratios, was directly proportional to the genetic relationship of the different species. The higher the Ka/Ks ratios, the longer the distance was. Moreover, 31 positively selected genes (PSGs) involved in carbohydrate metabolism and phenolic metabolism were identified and isolated, and nearly all PSGs except the MATE gene had a high expression in the DK or DKV species. It was hypothesized that these genes might contribute to the domestication of the DK species. Finally, we developed the expressed sequence tag-simple sequence repeat (EST-SSR) and identified 2 unique amplicons DKSSR10 and DKSSR39: the former was absent in the DC species but was present in the other species, the latter had a long amplification product in the DJ species.
Conclusion: This study presents the first transcriptome resources for the closely related species of persimmon and reveals interspecific genetic divergence. It is speculated that DK is derived from the hybridization of DD and DO species. Furthermore, our analysis suggests candidate PSGs that may be crucial for the adaptation, domestication, and speciation of persimmon relatives and suggests that DKSSR10 and DKSSSR39 could potentially serve as species-specific molecular markers.
Keywords: Comparative transcriptome; Diospyros; Domestication genes; Genetic divergence; Species.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Short communication: development and characterization of novel transcriptome-derived microsatellites for genetic analysis of persimmon.Genet Mol Res. 2014 Apr 16;13(2):3013-24. doi: 10.4238/2014.April.16.10. Genet Mol Res. 2014. PMID: 24782136
-
Characterization and comparison of EST-SSR and TRAP markers for genetic analysis of the Japanese persimmon Diospyros kaki.Genet Mol Res. 2013 Jan 9;12(3):2841-51. doi: 10.4238/2013.January.9.3. Genet Mol Res. 2013. PMID: 23359055
-
Development of simple sequence repeat markers in persimmon (Diospyros L.) and their potential use in related species.Genet Mol Res. 2015 Jan 30;14(1):609-18. doi: 10.4238/2015.January.30.2. Genet Mol Res. 2015. PMID: 25729996
-
Characterization of the teosinte transcriptome reveals adaptive sequence divergence during maize domestication.Mol Ecol Resour. 2016 Nov;16(6):1465-1477. doi: 10.1111/1755-0998.12526. Epub 2016 Mar 31. Mol Ecol Resour. 2016. PMID: 26990495
-
Interspecific chloroplast genome sequence diversity and genomic resources in Diospyros.BMC Plant Biol. 2018 Sep 26;18(1):210. doi: 10.1186/s12870-018-1421-3. BMC Plant Biol. 2018. PMID: 30257644 Free PMC article.
Cited by
-
Haplotype-resolved and chromosome-level reference genome assembly of Diospyros deyangensis provides insights into the evolution and juvenile growth of persimmon.Hortic Res. 2025 Jan 8;12(4):uhaf001. doi: 10.1093/hr/uhaf001. eCollection 2025 Apr. Hortic Res. 2025. PMID: 40078717 Free PMC article.
-
Exploring genetic diversity and population structure in Cinnamomum cassia (L.) J.Presl germplasm in China through phenotypic, chemical component, and molecular marker analyses.Front Plant Sci. 2024 Jul 3;15:1374648. doi: 10.3389/fpls.2024.1374648. eCollection 2024. Front Plant Sci. 2024. PMID: 39055357 Free PMC article.
-
Phylotranscriptomic discordance is best explained by incomplete lineage sorting within Allium subgenus Cyathophora and thus hemiplasy accounts for interspecific trait transition.Plant Divers. 2023 Jul 17;46(1):28-38. doi: 10.1016/j.pld.2023.07.004. eCollection 2024 Jan. Plant Divers. 2023. PMID: 38343588 Free PMC article.
-
Phenotypic, chemical component and molecular assessment of genetic diversity and population structure of Morinda officinalis germplasm.BMC Genomics. 2022 Aug 19;23(1):605. doi: 10.1186/s12864-022-08817-w. BMC Genomics. 2022. PMID: 35986256 Free PMC article.
-
Investigating phenotypic relationships in persimmon accessions through integrated proteomic and metabolomic analysis of corresponding fruits.Front Plant Sci. 2023 Jan 30;14:1093074. doi: 10.3389/fpls.2023.1093074. eCollection 2023. Front Plant Sci. 2023. PMID: 36794209 Free PMC article.
References
-
- Duangjai S, Samuel R, Munzinger J, Forest F, Wallnöfer B, Barfuss MHJ, et al. A multi-locus plastid phylogenetic analysis of the pantropical genus Diospyros (Ebenaceae), with an emphasis on the radiation and biogeographic origins of the new Caledonian endemic species. Mol Phylogenet Evol. 2009;52:602–620. doi: 10.1016/j.ympev.2009.04.021. - DOI - PubMed
-
- Lee S, Michael GG, Frank W. Ebenaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Beijing, Missouri, St. Louis: Science press, botanical garden press; 1996. pp. 215–234.
-
- Wallnöfer B. The biology and systematics of Ebenaceae: a review. Annalen des Naturhistorischen Museums in Wien Serie B für Botanik und Zoologie. 2001;103:485–512.
-
- Du XY, Zhang QL, Luo ZR. Comparison of four molecular markers for genetic analysis in Diospyros L. (Ebenaceae) Plant Syst Evol. 2009;281:171–181. doi: 10.1007/s00606-009-0199-z. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

