Scribble, Erbin, and Lano redundantly regulate epithelial polarity and apical adhesion complex
- PMID: 31147384
- PMCID: PMC6605793
- DOI: 10.1083/jcb.201804201
Scribble, Erbin, and Lano redundantly regulate epithelial polarity and apical adhesion complex
Abstract
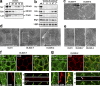
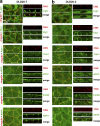
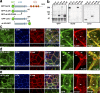
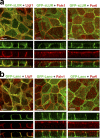
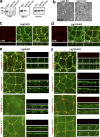
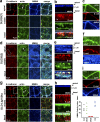
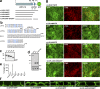
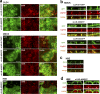
The basolateral protein Scribble (Scrib), a member of the LAP protein family, is essential for epithelial apicobasal polarity (ABP) in Drosophila However, a conserved function for this protein in mammals is unclear. Here we show that the crucial role for Scrib in ABP has remained obscure due to the compensatory function of two other LAP proteins, Erbin and Lano. A combined Scrib/Erbin/Lano knockout disorganizes the cell-cell junctions and the cytoskeleton. It also results in mislocalization of several apical (Par6, aPKC, and Pals1) and basolateral (Llgl1 and Llgl2) identity proteins. These defects can be rescued by the conserved "LU" region of these LAP proteins. Structure-function analysis of this region determined that the so-called LAPSDb domain is essential for basolateral targeting of these proteins, while the LAPSDa domain is essential for supporting the membrane basolateral identity and binding to Llgl. In contrast to the key role in Drosophila, mislocalization of Llgl proteins does not appear to be critical in the scrib ABP phenotype.
© 2019 Choi et al.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

