Insights into IgM-mediated complement activation based on in situ structures of IgM-C1-C4b
- PMID: 31147461
- PMCID: PMC6575175
- DOI: 10.1073/pnas.1901841116
Insights into IgM-mediated complement activation based on in situ structures of IgM-C1-C4b
Abstract
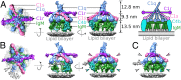
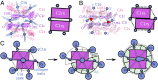
Antigen binding by serum Ig-M (IgM) protects against microbial infections and helps to prevent autoimmunity, but causes life-threatening diseases when mistargeted. How antigen-bound IgM activates complement-immune responses remains unclear. We present cryoelectron tomography structures of IgM, C1, and C4b complexes formed on antigen-bearing lipid membranes by normal human serum at 4 °C. The IgM-C1-C4b complexes revealed C4b product release as the temperature-limiting step in complement activation. Both IgM hexamers and pentamers adopted hexagonal, dome-shaped structures with Fab pairs, dimerized by hinge domains, bound to surface antigens that support a platform of Fc regions. C1 binds IgM through widely spread C1q-collagen helices, with C1r proteases pointing outward and C1s bending downward and interacting with surface-attached C4b, which further interacts with the adjacent IgM-Fab2 and globular C1q-recognition unit. Based on these data, we present mechanistic models for antibody-mediated, C1q-transmitted activation of C1 and for C4b deposition, while further conformational rearrangements are required to form C3 convertases.
Keywords: C1; IgM; complement; cryoelectron tomography; subtomogram averaging.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

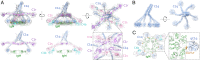

References
-
- Boes M., Role of natural and immune IgM antibodies in immune responses. Mol. Immunol. 37, 1141–1149 (2000). - PubMed
-
- Mäkelä O., Ruoslahti E., Seppälä I. J., Affinity of IgM and IgG antibodies. Immunochemistry 7, 917–932 (1970). - PubMed
-
- Quartier P., Potter P. K., Ehrenstein M. R., Walport M. J., Botto M., Predominant role of IgM-dependent activation of the classical pathway in the clearance of dying cells by murine bone marrow-derived macrophages in vitro. Eur. J. Immunol. 35, 252–260 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

