Linking the resistome and plasmidome to the microbiome
- PMID: 31147603
- PMCID: PMC6776055
- DOI: 10.1038/s41396-019-0446-4
Linking the resistome and plasmidome to the microbiome
Abstract
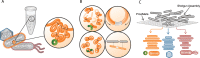
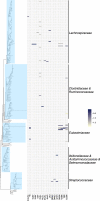
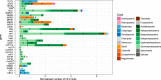
The rapid spread of antibiotic resistance among bacterial pathogens is a serious human health threat. While a range of environments have been identified as reservoirs of antibiotic resistance genes (ARGs), we lack understanding of the origins of these ARGs and their spread from environment to clinic. This is partly due to our inability to identify the natural bacterial hosts of ARGs and the mobile genetic elements that mediate this spread, such as plasmids and integrons. Here we demonstrate that the in vivo proximity-ligation method Hi-C can reconstruct a known plasmid-host association from a wastewater community, and identify the in situ host range of ARGs, plasmids, and integrons by physically linking them to their host chromosomes. Hi-C detected both previously known and novel associations between ARGs, mobile genetic elements and host genomes, thus validating this method. We showed that IncQ plasmids and class 1 integrons had the broadest host range in this wastewater, and identified bacteria belonging to Moraxellaceae, Bacteroides, and Prevotella, and especially Aeromonadaceae as the most likely reservoirs of ARGs in this community. A better identification of the natural carriers of ARGs will aid the development of strategies to limit resistance spread to pathogens.
Conflict of interest statement
MP, SS, and IL are employees and shareholders of Phase Genomics, Inc—a company commercializing proximity-ligation technology. IL and SS are executives at Phase Genomics, Inc.
Figures


References
-
- Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, Saeed DK, et al. Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. MBio. 2018;9:e00105–18. doi: 10.1128/mBio.00105-18. - DOI - PMC - PubMed
-
- Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–61. doi: 10.1128/AAC.45.4.1151-1161.2001. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

