LnCompare: gene set feature analysis for human long non-coding RNAs
- PMID: 31147707
- PMCID: PMC6602513
- DOI: 10.1093/nar/gkz410
LnCompare: gene set feature analysis for human long non-coding RNAs
Abstract
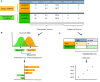
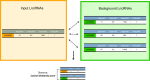
Interest in the biological roles of long noncoding RNAs (lncRNAs) has resulted in growing numbers of studies that produce large sets of candidate genes, for example, differentially expressed between two conditions. For sets of protein-coding genes, ontology and pathway analyses are powerful tools for generating new insights from statistical enrichment of gene features. Here we present the LnCompare web server, an equivalent resource for studying the properties of lncRNA gene sets. The Gene Set Feature Comparison mode tests for enrichment amongst a panel of quantitative and categorical features, spanning gene structure, evolutionary conservation, expression, subcellular localization, repetitive sequences and disease association. Moreover, in Similar Gene Identification mode, users may identify other lncRNAs by similarity across a defined range of features. Comprehensive results may be downloaded in tabular and graphical formats, in addition to the entire feature resource. LnCompare will empower researchers to extract useful hypotheses and candidates from lncRNA gene sets.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Bonetti A., Carninci P.. From bench to bedside: the long journey of long non-coding RNAs. Curr. Opin. Syst. Biol. 2017; 3:119–124.

