Pichia pastoris protease-deficient and auxotrophic strains generated by a novel, user-friendly vector toolbox for gene deletion
- PMID: 31148217
- PMCID: PMC6771850
- DOI: 10.1002/yea.3426
Pichia pastoris protease-deficient and auxotrophic strains generated by a novel, user-friendly vector toolbox for gene deletion
Abstract
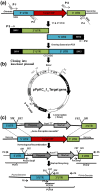
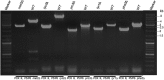
Targeted gene knockouts play an important role in the study of gene function. For the generation of knockouts in the industrially important yeast Pichia pastoris, several protocols have been published to date. Nevertheless, creating a targeted knockout in P. pastoris still is a time-consuming process, as the existing protocols are labour intensive and/or prone to accumulate nucleotide mutations. In this study, we introduce a novel, user-friendly vector-based system for the generation of targeted knockouts in P. pastoris. Upon confirming the successful knockout, respective selection markers can easily be recycled. Excision of the marker is mediated by Flippase (Flp) recombinase and occurs at high frequency (≥95%). We validated our knockout system by deleting 20 (confirmed and putative) protease genes and five genes involved in biosynthetic pathways. For the first time, we describe gene deletions of PRO3 and PHA2 in P. pastoris, genes involved in proline, and phenylalanine biosynthesis, respectively. Unexpectedly, knockout strains of PHA2 did not display the anticipated auxotrophy for phenylalanine but rather showed a bradytroph phenotype on minimal medium hinting at an alternative but less efficient pathway for production of phenylalanine exists in P. pastoris. Overall, all knockout vectors can easily be adapted to the gene of interest and strain background by efficient exchange of target homology regions and selection markers in single cloning steps. Average knockout efficiencies for all 25 genes were shown to be 40%, which is comparably high.
Keywords: P. pastoris; auxotrophic strains; gene disruption; knockout plasmids; proteases-deficient strains.
© 2019 The Authors Yeast Published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Austin, R. J. , Kuestner, R. E. , Chang, D. K. , Madden, K. R. , & Martin, D. B. (2011). SILAC compatible strain of Pichia pastoris for expression of isotopically labeled protein standards and quantitative proteomics. Journal of Proteome Research, 10, 5251–5259. 10.1021/pr200551e - DOI - PMC - PubMed
-
- Brachmann, C. B. , Davies, A. , Cost, G. J. , Caputo, E. , Li, J. , Hieter, P. , & Boeke, J. D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR‐mediated gene disruption and other applications. Yeast, 14, 115–132. 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
