Mapping connections in signaling networks with ambiguous modularity
- PMID: 31149348
- PMCID: PMC6533310
- DOI: 10.1038/s41540-019-0096-1
Mapping connections in signaling networks with ambiguous modularity
Abstract
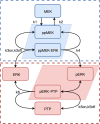
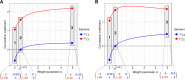
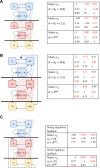
Modular Response Analysis (MRA) is a suite of methods that under certain assumptions permits the precise reconstruction of both the directions and strengths of connections between network modules from network responses to perturbations. Standard MRA assumes that modules are insulated, thereby neglecting the existence of inter-modular protein complexes. Such complexes sequester proteins from different modules and propagate perturbations to the protein abundance of a downstream module retroactively to an upstream module. MRA-based network reconstruction detects retroactive, sequestration-induced connections when an enzyme from one module is substantially sequestered by its substrate that belongs to a different module. Moreover, inferred networks may surprisingly depend on the choice of protein abundances that are experimentally perturbed, and also some inferred connections might be false. Here, we extend MRA by introducing a combined computational and experimental approach, which allows for a computational restoration of modular insulation, unmistakable network reconstruction and discrimination between solely regulatory and sequestration-induced connections for a range of signaling pathways. Although not universal, our approach extends MRA methods to signaling networks with retroactive interactions between modules arising from enzyme sequestration effects.
Keywords: Applied mathematics; Biochemical networks; Computer modelling.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

