The genomes of polyextremophilic cyanidiales contain 1% horizontally transferred genes with diverse adaptive functions
- PMID: 31149898
- PMCID: PMC6629376
- DOI: 10.7554/eLife.45017
The genomes of polyextremophilic cyanidiales contain 1% horizontally transferred genes with diverse adaptive functions
Abstract
The role and extent of horizontal gene transfer (HGT) in eukaryotes are hotly disputed topics that impact our understanding of the origin of metabolic processes and the role of organelles in cellular evolution. We addressed this issue by analyzing 10 novel Cyanidiales genomes and determined that 1% of their gene inventory is HGT-derived. Numerous HGT candidates share a close phylogenetic relationship with prokaryotes that live in similar habitats as the Cyanidiales and encode functions related to polyextremophily. HGT candidates differ from native genes in GC-content, number of splice sites, and gene expression. HGT candidates are more prone to loss, which may explain the absence of a eukaryotic pan-genome. Therefore, the lack of a pan-genome and cumulative effects fail to provide substantive arguments against our hypothesis of recurring HGT followed by differential loss in eukaryotes. The maintenance of 1% HGTs, even under selection for genome reduction, underlines the importance of non-endosymbiosis related foreign gene acquisition.
Keywords: Cyanidiales; evolution; evolutionary biology; genome; horizontal gene transfer; lateral gene transfer; red algae.
© 2019, Rossoni et al.
Conflict of interest statement
AR, DP, MS, DL, PL, DB, AW No competing interests declared
Figures





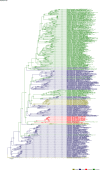
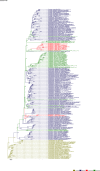

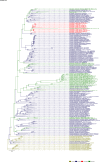
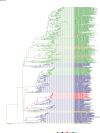
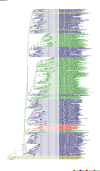
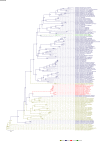
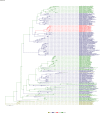
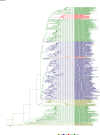


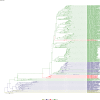
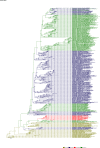

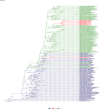
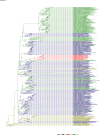

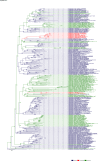
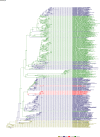
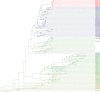
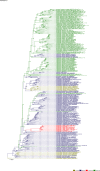
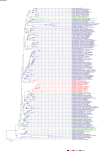
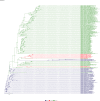
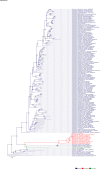
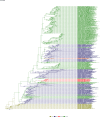
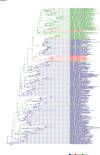
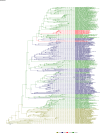
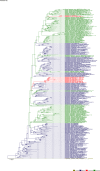
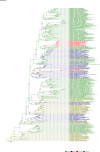
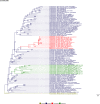



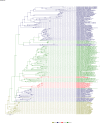
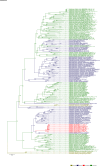
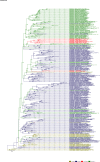

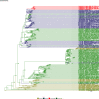


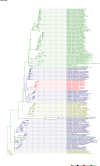
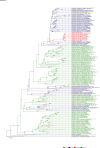

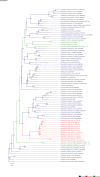
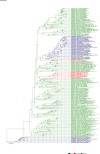
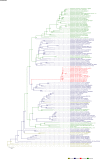
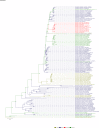
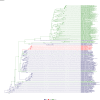

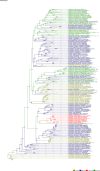
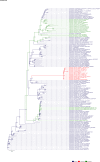

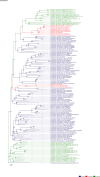

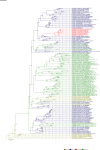
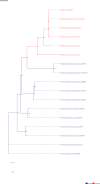

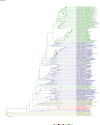

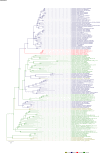

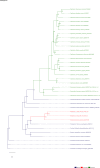
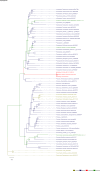
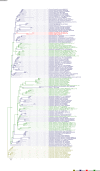


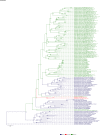


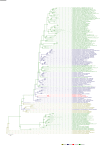
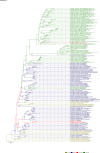

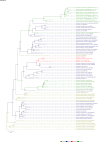
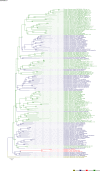


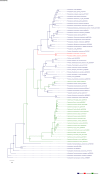
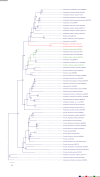

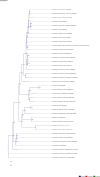

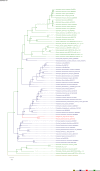

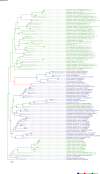






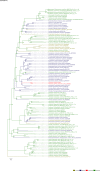



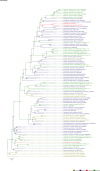

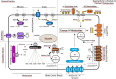
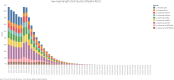

















Comment in
-
Adapting for life in the extreme.Elife. 2019 Jul 15;8:e48999. doi: 10.7554/eLife.48999. Elife. 2019. PMID: 31305242 Free PMC article.
References
-
- Aiuppa A. The aquatic geochemistry of arsenic in volcanic groundwaters from southern italy. Science. 2003;18:1283–1296. doi: 10.1016/S0883-2927(03)00051-9. - DOI
-
- Albertano P. The taxonomic position of cyanidium, cyanidioschyzon and galdieria: an update. Hydrobiologia. 2000;433:137–143. doi: 10.1023/A:1004031123806. - DOI
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. the gene ontology consortium. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

