In-Vivo and Ex-Vivo Tissue Analysis through Hyperspectral Imaging Techniques: Revealing the Invisible Features of Cancer
- PMID: 31151223
- PMCID: PMC6627361
- DOI: 10.3390/cancers11060756
In-Vivo and Ex-Vivo Tissue Analysis through Hyperspectral Imaging Techniques: Revealing the Invisible Features of Cancer
Abstract
In contrast to conventional optical imaging modalities, hyperspectral imaging (HSI) is able to capture much more information from a certain scene, both within and beyond the visual spectral range (from 400 to 700 nm). This imaging modality is based on the principle that each material provides different responses to light reflection, absorption, and scattering across the electromagnetic spectrum. Due to these properties, it is possible to differentiate and identify the different materials/substances presented in a certain scene by their spectral signature. Over the last two decades, HSI has demonstrated potential to become a powerful tool to study and identify several diseases in the medical field, being a non-contact, non-ionizing, and a label-free imaging modality. In this review, the use of HSI as an imaging tool for the analysis and detection of cancer is presented. The basic concepts related to this technology are detailed. The most relevant, state-of-the-art studies that can be found in the literature using HSI for cancer analysis are presented and summarized, both in-vivo and ex-vivo. Lastly, we discuss the current limitations of this technology in the field of cancer detection, together with some insights into possible future steps in the improvement of this technology.
Keywords: artificial intelligence; biomedical optical imaging; cancer; clinical diagnosis; hyperspectral imaging; machine learning; medical diagnostic imaging.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


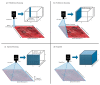
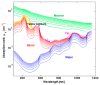


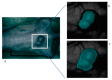
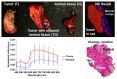
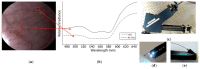


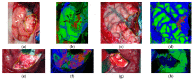
References
-
- Kamruzzaman M., Sun D.W. Computer Vision Technology for Food Quality Evaluation. Elsevier; Amsterdam, The Netherlands: 2016. Introduction to Hyperspectral Imaging Technology; pp. 111–139. - DOI
-
- Starr C., Evers C., Starr L. Biology: Concepts and Applications without Physiology. Cengage Learning; Boston, MA, USA: 2010.
-
- Manolakis D., Shaw G. Detection Algorithms for Hyperspectral Imaging Applications. IEEE Signal Process. Mag. 2002;19:29–43. doi: 10.1109/79.974724. - DOI
-
- Govender M., Chetty K., Bulcock H. A review of hyperspectral remote sensing and its application in vegetation and water resource studies. Water SA. 2009;33:145–152. doi: 10.4314/wsa.v33i2.49049. - DOI
-
- Van der Meer F.D., van der Werff H.M.A., van Ruitenbeek F.J.A., Hecker C.A., Bakker W.H., Noomen M.F., van der Meijde M., Carranza E.J.M., de Smeth J.B., Woldai T. Multi- and hyperspectral geologic remote sensing: A review. Int. J. Appl. Earth Obs. Geoinform. 2012;14:112–128. doi: 10.1016/j.jag.2011.08.002. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

