Arc Oligomerization Is Regulated by CaMKII Phosphorylation of the GAG Domain: An Essential Mechanism for Plasticity and Memory Formation
- PMID: 31151856
- PMCID: PMC6625847
- DOI: 10.1016/j.molcel.2019.05.004
Arc Oligomerization Is Regulated by CaMKII Phosphorylation of the GAG Domain: An Essential Mechanism for Plasticity and Memory Formation
Abstract
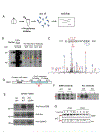
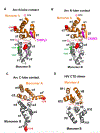
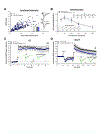
Arc is a synaptic protein essential for memory consolidation. Recent studies indicate that Arc originates in evolution from a Ty3-Gypsy retrotransposon GAG domain. The N-lobe of Arc GAG domain acquired a hydrophobic binding pocket in higher vertebrates that is essential for Arc's canonical function to weaken excitatory synapses. Here, we report that Arc GAG also acquired phosphorylation sites that can acutely regulate its synaptic function. CaMKII phosphorylates the N-lobe of the Arc GAG domain and disrupts an interaction surface essential for high-order oligomerization. In Purkinje neurons, CaMKII phosphorylation acutely reverses Arc's synaptic action. Mutant Arc that cannot be phosphorylated by CaMKII enhances metabotropic receptor-dependent depression in the hippocampus but does not alter baseline synaptic transmission or long-term potentiation. Behavioral studies indicate that hippocampus- and amygdala-dependent learning requires Arc GAG domain phosphorylation. These studies provide an atomic model for dynamic and local control of Arc function underlying synaptic plasticity and memory.
Keywords: Arc; CAMKII; capsid; evolution; learning and memory; phosphorylation; polymerization; synapse plasticity.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures




Similar articles
-
The Capsid Domain of Arc Changes Its Oligomerization Propensity through Direct Interaction with the NMDA Receptor.Structure. 2019 Jul 2;27(7):1071-1081.e5. doi: 10.1016/j.str.2019.04.001. Epub 2019 May 9. Structure. 2019. PMID: 31080121
-
Structural basis of arc binding to synaptic proteins: implications for cognitive disease.Neuron. 2015 Apr 22;86(2):490-500. doi: 10.1016/j.neuron.2015.03.030. Epub 2015 Apr 9. Neuron. 2015. PMID: 25864631 Free PMC article.
-
Spatial exploration induces ARC, a plasticity-related immediate-early gene, only in calcium/calmodulin-dependent protein kinase II-positive principal excitatory and inhibitory neurons of the rat forebrain.J Comp Neurol. 2006 Sep 20;498(3):317-29. doi: 10.1002/cne.21003. J Comp Neurol. 2006. PMID: 16871537
-
Arc - An endogenous neuronal retrovirus?Semin Cell Dev Biol. 2018 May;77:73-78. doi: 10.1016/j.semcdb.2017.09.029. Epub 2017 Sep 24. Semin Cell Dev Biol. 2018. PMID: 28941877 Free PMC article. Review.
-
CaMKII regulation in information processing and storage.Trends Neurosci. 2012 Oct;35(10):607-18. doi: 10.1016/j.tins.2012.05.003. Epub 2012 Jun 19. Trends Neurosci. 2012. PMID: 22717267 Free PMC article. Review.
Cited by
-
Analysis of personality traits and academic performance in higher education at a Colombian university.Heliyon. 2021 May 11;7(5):e06998. doi: 10.1016/j.heliyon.2021.e06998. eCollection 2021 May. Heliyon. 2021. PMID: 34036192 Free PMC article.
-
Development and Validation of Arc Nanobodies: New Tools for Probing Arc Dynamics and Function.Neurochem Res. 2022 Sep;47(9):2656-2666. doi: 10.1007/s11064-022-03573-5. Epub 2022 Mar 20. Neurochem Res. 2022. PMID: 35307777 Free PMC article.
-
High-affinity anti-Arc nanobodies provide tools for structural and functional studies.PLoS One. 2022 Jun 7;17(6):e0269281. doi: 10.1371/journal.pone.0269281. eCollection 2022. PLoS One. 2022. PMID: 35671319 Free PMC article.
-
Loss of GDE2 leads to complex behavioral changes including memory impairment.Behav Brain Funct. 2024 Apr 4;20(1):7. doi: 10.1186/s12993-024-00234-1. Behav Brain Funct. 2024. PMID: 38575965 Free PMC article.
-
Molecular physiology of Arc/Arg3.1: The oligomeric state hypothesis of synaptic plasticity.Acta Physiol (Oxf). 2022 Nov;236(3):e13886. doi: 10.1111/apha.13886. Epub 2022 Sep 20. Acta Physiol (Oxf). 2022. PMID: 36073248 Free PMC article. Review.
References
-
- Campillos M, Doerks T, Shah PK, and Bork P (2006). Computational characterization of multiple Gag-like human proteins. Trends Genet 22, 585–589. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

