Individual Physiological Adaptations Enable Selected Bacterial Taxa To Prevail during Long-Term Incubations
- PMID: 31152013
- PMCID: PMC6643244
- DOI: 10.1128/AEM.00825-19
Individual Physiological Adaptations Enable Selected Bacterial Taxa To Prevail during Long-Term Incubations
Abstract
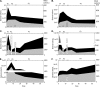
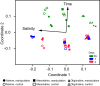
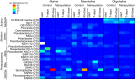
Enclosure experiments are frequently used to investigate the impact of changing environmental conditions on microbial assemblages. Yet, how the incubation itself challenges complex bacterial communities is thus far unknown. In this study, metaproteomic profiling, 16S rRNA gene analyses, and cell counts were combined to evaluate bacterial communities derived from marine, mesohaline, and oligohaline conditions after long-term batch incubations. Early in the experiment, the three bacterial communities were highly diverse and differed significantly in their compositions. Manipulation of the enclosures with terrigenous dissolved organic carbon resulted in notable differences compared to the control enclosures at this early phase of the experiment. However, after 55 days, bacterial communities in the manipulated and the control enclosures under marine and mesohaline conditions were all dominated by gammaproteobacterium Spongiibacter In the oligohaline enclosures, actinobacterial cluster I of the hgc group (hgc-I) remained abundant in the late phase of the incubation. Metaproteome analyses suggested that the ability to use outer membrane-based internal energy stores, in addition to the previously described grazing resistance, may enable the gammaproteobacterium Spongiibacter to prevail in long-time incubations. Under oligohaline conditions, the utilization of external recalcitrant carbon appeared to be more important (hgc-I). Enclosure experiments with complex natural microbial communities are important tools to investigate the effects of manipulations. However, species-specific properties, such as individual carbon storage strategies, can cause manipulation-independent effects and need to be considered when interpreting results from enclosures.IMPORTANCE In microbial ecology, enclosure studies are often used to investigate the effect of single environmental factors on complex bacterial communities. However, in addition to the manipulation, unintended effects ("bottle effect") may occur due to the enclosure itself. In this study, we analyzed the bacterial communities that originated from three different salinities of the Baltic Sea, comparing their compositions and physiological activities both at the early stage and after 55 days of incubation. Our results suggested that internal carbon storage strategies impact the success of certain bacterial species, independent of the experimental manipulation. Thus, while enclosure experiments remain valid tools in environmental research, microbial community composition shifts must be critically followed. This investigation of the metaproteome during long-term batch enclosures expanded our current understanding of the so-called "bottle effect," which is well known to occur during enclosure experiments.
Keywords: Baltic Sea; Spongiibacter; bottle effect; enclosure; salinity.
Copyright © 2019 American Society for Microbiology.
Figures



Similar articles
-
Differential responses of marine, mesohaline and oligohaline bacterial communities to the addition of terrigenous carbon.Environ Microbiol. 2017 Aug;19(8):3098-3117. doi: 10.1111/1462-2920.13784. Epub 2017 May 29. Environ Microbiol. 2017. PMID: 28474480
-
Benthic Bacterial Community Composition in the Oligohaline-Marine Transition of Surface Sediments in the Baltic Sea Based on rRNA Analysis.Front Microbiol. 2018 Feb 19;9:236. doi: 10.3389/fmicb.2018.00236. eCollection 2018. Front Microbiol. 2018. PMID: 29520255 Free PMC article.
-
Responses of Baltic Sea ice and open-water natural bacterial communities to salinity change.Appl Environ Microbiol. 2005 Aug;71(8):4364-71. doi: 10.1128/AEM.71.8.4364-4371.2005. Appl Environ Microbiol. 2005. PMID: 16085826 Free PMC article.
-
Diversity and abundance of "Pelagibacterales" (SAR11) in the Baltic Sea salinity gradient.Syst Appl Microbiol. 2014 Dec;37(8):601-4. doi: 10.1016/j.syapm.2014.09.002. Epub 2014 Oct 5. Syst Appl Microbiol. 2014. PMID: 25444644
-
Review of the Effects of Enclosure Complexity and Design on the Behaviour and Physiology of Zoo Animals.Animals (Basel). 2023 Apr 7;13(8):1277. doi: 10.3390/ani13081277. Animals (Basel). 2023. PMID: 37106840 Free PMC article. Review.
Cited by
-
Distinct stages of the intestinal bacterial community of Ampullaceana balthica after salinization.Front Microbiol. 2022 Aug 30;13:767334. doi: 10.3389/fmicb.2022.767334. eCollection 2022. Front Microbiol. 2022. PMID: 36110301 Free PMC article.
References
-
- Duarte CM, Gasol JM, Vaqué D. 1997. Role of experimental approaches in marine microbial ecology. Aquat Microb Ecol 13:101–111. doi:10.3354/ame013101. - DOI
-
- Massana R, Jürgens K. 2003. Composition and population dynamics of planktonic bacteria and bacterivorous flagellates in seawater chemostat cultures. Aquat Microb Ecol 32:11–22. doi:10.3354/ame032011. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

