Giant group I intron in a mitochondrial genome is removed by RNA back-splicing
- PMID: 31153363
- PMCID: PMC6545197
- DOI: 10.1186/s12867-019-0134-y
Giant group I intron in a mitochondrial genome is removed by RNA back-splicing
Abstract
Background: The mitochondrial genomes of mushroom corals (Corallimorpharia) are remarkable for harboring two complex group I introns; ND5-717 and COI-884. How these autocatalytic RNA elements interfere with mitochondrial RNA processing is currently not known. Here, we report experimental support for unconventional processing events of ND5-717 containing RNA.
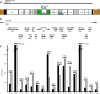
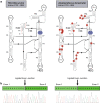
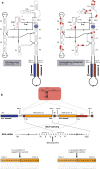
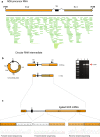
Results: We obtained the complete mitochondrial genome sequences and corresponding mitochondrial transcriptomes of the two distantly related corallimorpharian species Ricordea yuma and Amplexidiscus fenestrafer. All mitochondrial genes were found to be expressed at the RNA-level. Both introns were perfectly removed by autocatalytic splicing, but COI-884 excision appeared more efficient than ND5-717. ND5-717 was organized into giant group I intron elements of 18.1 kb and 19.3 kb in A. fenestrafer and R. yuma, respectively. The intron harbored almost the entire mitochondrial genome embedded within the P8 peripheral segment.
Conclusion: ND5-717 was removed by group I intron splicing from a small primary transcript that contained a permutated intron-exon arrangement. The splicing pathway involved a circular exon-containing RNA intermediate, which is a hallmark of RNA back-splicing. ND5-717 represents the first reported natural group I intron that becomes excised by back-splicing from a permuted precursor RNA. Back-splicing may explain why Corallimorpharia mitochondrial genomes tolerate giant group I introns.
Keywords: Amplexidiscus; Back-splicing; Catalytic RNA; Group I intron; Intron retention; Mitochondrial RNA; Ribozyme; Ricordea.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

