circRNA_0025202 Regulates Tamoxifen Sensitivity and Tumor Progression via Regulating the miR-182-5p/FOXO3a Axis in Breast Cancer
- PMID: 31153828
- PMCID: PMC6731174
- DOI: 10.1016/j.ymthe.2019.05.011
circRNA_0025202 Regulates Tamoxifen Sensitivity and Tumor Progression via Regulating the miR-182-5p/FOXO3a Axis in Breast Cancer
Erratum in
-
circRNA_0025202 Regulates Tamoxifen Sensitivity and Tumor Progression via Regulating the miR-182-5p/FOXO3a Axis in Breast Cancer.Mol Ther. 2021 Dec 1;29(12):3525-3527. doi: 10.1016/j.ymthe.2021.11.002. Epub 2021 Nov 12. Mol Ther. 2021. PMID: 34774125 Free PMC article. No abstract available.
Abstract
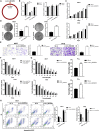
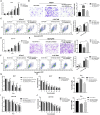
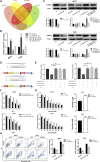
Tamoxifen is the most commonly used endocrine therapy for patients with hormone receptor (HR)-positive breast cancer. Despite its initial therapeutic efficacy, many patients eventually develop drug resistance, which remains a serious clinical challenge. To investigate roles of circular RNAs (circRNAs) in tamoxifen resistance, a tamoxifen-resistant MCF-7 cell line was established and screened for its circRNA expression profile by RNA sequencing. hsa_circ_0025202, a circRNA that was significantly downregulated, was selected for further investigation. Using a large cohort of clinical specimens, we found that hsa_circ_0025202 exhibited low expression in cancer tissues and was negatively correlated with lymphatic metastasis and histological grade. Gain- and loss-of-function assays indicated that hsa_circ_0025202 could inhibit cell proliferation, colony formation, and migration and increase cell apoptosis and sensitivity to tamoxifen. Bioinformatics and luciferase reporter assays verified that hsa_circ_0025202 could act as a miRNA sponge for miR-182-5p and further regulate the expression and activity of FOXO3a. Functional studies revealed that tumor inhibition and tamoxifen sensitization effects of hsa_circ_0025202 were achieved via the miR-182-5p/FOXO3a axis. Moreover, in vivo experiments confirmed that hsa_circ_0025202 could suppress tumor growth and enhance tamoxifen efficacy. Taken together, hsa_circ_0025202 served an anti-oncogenic role in HR-positive breast cancer, and it could be exploited as a novel marker for tamoxifen-resistant breast cancer.
Keywords: breast cancer; ceRNA; circRNA; hsa_circ_0025202; tamoxifen resistance.
Copyright © 2019 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. - PubMed
-
- Ojo D., Wei F., Liu Y., Wang E., Zhang H., Lin X., Wong N., Bane A., Tang D. Factors Promoting Tamoxifen Resistance in Breast Cancer via Stimulating Breast Cancer Stem Cell Expansion. Curr. Med. Chem. 2015;22:2360–2374. - PubMed
-
- Tryfonidis K., Zardavas D., Katzenellenbogen B.S., Piccart M. Endocrine treatment in breast cancer: Cure, resistance and beyond. Cancer Treat. Rev. 2016;50:68–81. - PubMed
-
- Osborne C.K., Schiff R., Fuqua S.A., Shou J. Estrogen receptor: current understanding of its activation and modulation. Clin. Cancer Res. 2001;7(Suppl 12):4338s–4342s. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

