The Mechanisms of Substrate Selection, Catalysis, and Translocation by the Elongating RNA Polymerase
- PMID: 31153902
- PMCID: PMC6874739
- DOI: 10.1016/j.jmb.2019.05.042
The Mechanisms of Substrate Selection, Catalysis, and Translocation by the Elongating RNA Polymerase
Abstract
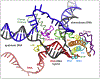
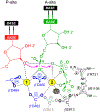
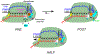
Multi-subunit DNA-dependent RNA polymerases synthesize all classes of cellular RNAs, ranging from short regulatory transcripts to gigantic messenger RNAs. RNA polymerase has to make each RNA product in just one try, even if it takes millions of successive nucleotide addition steps. During each step, RNA polymerase selects a correct substrate, adds it to a growing chain, and moves one nucleotide forward before repeating the cycle. However, RNA synthesis is anything but monotonous: RNA polymerase frequently pauses upon encountering mechanical, chemical and torsional barriers, sometimes stepping back and cleaving off nucleotides from the growing RNA chain. A picture in which these intermittent dynamics enable processive, accurate, and controllable RNA synthesis is emerging from complementary structural, biochemical, computational, and single-molecule studies. Here, we summarize our current understanding of the mechanism and regulation of the on-pathway transcription elongation. We review the details of substrate selection, catalysis, proofreading, and translocation, focusing on rate-limiting steps, structural elements that modulate them, and accessory proteins that appear to control RNA polymerase translocation.
Keywords: RNA polymerase; proofreading; transcription elongation; translocation; trigger loop.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Figures

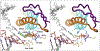


References
-
- Hurwitz J The discovery of RNA polymerase. J Biol Chem. 2005;280:42477–85. - PubMed
-
- Lee F, Squires CL, Squires C, Yanofsky C. Termination of transcription in vitro in the Escherichia coli tryptophan operon leader region. J Mol Biol. 1976;103:383–93. - PubMed
-
- Kingston RE, Chamberlin MJ. Pausing and attenuation of in vitro transcription in the rrnB operon of E. coli. Cell. 1981;27:523–31. - PubMed
-
- Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

